WRF/DART Tutorial Materials for the Manhattan Release.
Introduction
This document will describe how to get started with your own Weather Research and Forecasting (WRF) data assimilation experiments using DART and only covers the WRF-specific aspects of coupling with DART. It is not wise to try to run WRF-DART if you have no experience with either WRF or DART.
This tutorial was designed to be compatible with WRF V3.9.1 and was tested with DART V11.0.2. Other releases of WRF may or may not be backwards or forwards compatible with this tutorial. Prior to running this tutorial, we urge the users to familarize themselves with the WRF system (WRF_ARW, WPS and WRFDA), and to read through the WRFv3.9 User’s Guide and the WRF model tutorials
The DART team is not responsible for and does not maintain the WRF code. For WRF related issues check out the WRF User Forum or the WRF github page.
If you are new to DART, we recommend that you become familiar with DART by working through the DART Tutorial and then understanding the DART getting started documentation.
This tutorial is not a toy simulation, but represents a realistic WRF-DART assimilation for the continental United States. It uses a WRF ensemble of 50 members that will be initialized from GFS initial conditions at 2017/04/27 00:00 UTC. The data included in the tutorial lasts until 2017/04/30 18:00 UTC. During this period, there was a strong rain and wind event that affected a large portion of the United States, causing record rains, localized flooding, and numerous tornadoes. For more information on the physical account of this case, see weather.gov.
By default, the tutorial case will only cover 12 hours of this event starting at 2017/04/27 00:00 UTC. The WRF model will be “spun-up” for six hours to generate a prior distribution. An assimilation of PREPBUFR observations will then be performed at 06:00 UTC, at which time analysis files will be generated to begin a new ensemble forecast. The WRF model will be advanced for 6 hours and a final assimilation cycle will be performed at 12:00 UTC. This process could then continue in order to investigate the strong rain and wind event. On NSF NCAR’s Derecho, the tutorial requires at least 30 minutes of run time, and can take much longer (1-2 hours) depending upon the PBS queue wait time.
The goal of this tutorial is to demonstrate how WRF-DART works, and to provide an understanding of the major steps within a data assimilation (DA) experiment. However, you will need to do additional work before you can apply WRF-DART to your own research application, as some of the steps involved in this tutorial (in particular, the perturbation bank and the observation sequence files) are provided for you in order to simplify the process. We provide a diagnostic section at the end of the tutorial to assess the skill/success of the assimilation. Be aware, an assimilation is not successful just because it runs to completion. A successful assimilation generally uses the vast majority of the observations provided and minimizes the bias and RMSE between the posterior model state and the observations.
Finally, if you are not running on the NSF NCAR Derecho (PBS) supercomputing system, you will need to customize the assimilation scripts (located in /DART/models/wrf/shell_scripts/) to match the details of your particular system. Specifically, you will need to edit the DART csh scripting to match your system settings whether that be, for example, a PBS, SLURM or LSF HPC system. Although the DART team can offer advice on how to customize the scripting to accomodate your HPC system, your HPC system administrator is likely the best resource to resolve these issues.
Important
The tutorial scripting and instructions are based on the NSF NCAR supercomputer Derecho, so you will need to edit the scripts and interpret the instructions for other HPC systems. The scripting uses examples of a PBS queuing system (e.g. Derecho) and LSF queuing system (e.g. decommissioned Yellowstone). You can use these as a template for your own system.
Step 1: Setup
There are several required dependencies for the executables and WRF-DART scripting components. On NSF NCAR’s Derecho, users have reported success building WRF, WPS, WRFDA, and DART using gfortan with the following module environment. Note: not all modules listed below are a requirement to compile and run the tutorial.
Currently Loaded Modules: 1) ncarenv/23.09 (S) 3) udunits/2.2.28 5) ncarcompilers/1.0.0 7) cray-mpich/8.1.27 9) netcdf-mpi/4.9.2 2) gcc/12.2.0 4) ncview/2.1.9 6) craype/2.7.23 8) hdf5-mpi/1.12.2 10) hdf/4.2.15
In addition, you’ll need to load the nco and ncl modules to run the set of scripts that accompany the tutorial. For Derecho the nco and ncl packages can be automatically loaded using the following commands:
module load nco module load ncl/6.6.2
These commands are provided by default with the param.csh script. More details are provided below. There are multiple phases for the setup: building the DART executables, downloading the initial WRF boundary conditions, building (or using existing) WRF executables, and configuring and staging the scripting needed to perform an experiment.
Build the DART executables.
If you have not already, see Getting Started to download the DART software package. Set an environment variable DART_DIR to point to your base DART directory. How to do this will depend on which shell you are using.
shell |
command |
|---|---|
tcsh |
|
bash |
|
In either case, you will replace <path_to_your_dart_installation> with the actual path to your DART installation. If you are using another shell, refer to your shell-specific documentation on how to set an environment variable.
Building the DART executables for the tutorial follows the same process
as building any of the DART executables. Configure the mkmf.template
file for your system, configure the input.nml for the model you want
to compile, and run quickbuild.sh (which is not necessarily quick,
but it is quicker than doing it by hand) to compile all the programs you
might need for an experiment with that model.
It is assumed you have successfully configured the
$DART_DIR/build_templates/mkmf.templatefile for your system. If not, you will need to do so now. See Getting Started for more detail, if necessary.
Important
If using gfortan to compile DART on Derecho, a successful configuration
of the mkmf.template includes using the mkmf.template.gfortan script
and customizing the compiler flags as follows:
FFLAGS = -O2 -ffree-line-length-none -fallow-argument-mismatch -fallow-invalid-boz $(INCS)
[OPTIONAL] Modify the DART code to use 32bit reals. Most WRF/DART users run both the WRF model and the DART assimilation code using 32bit reals. This is not the default for the DART code. Make this single code change before building the DART executables to compile all reals as 32bit reals.
Edit
$DART_DIR/assimilation_code/modules/utilities/types_mod.f90with your favorite editor. Change! real precision: ! TO RUN WITH REDUCED PRECISION REALS (and use correspondingly less memory) ! comment OUT the r8 definition below and use the second one: integer, parameter :: r4 = SELECTED_REAL_KIND(6,30) integer, parameter :: r8 = SELECTED_REAL_KIND(12) ! 8 byte reals !integer, parameter :: r8 = r4 ! alias r8 to r4
to
! real precision: ! TO RUN WITH REDUCED PRECISION REALS (and use correspondingly less memory) ! comment OUT the r8 definition below and use the second one: integer, parameter :: r4 = SELECTED_REAL_KIND(6,30) ! integer, parameter :: r8 = SELECTED_REAL_KIND(12) ! 8 byte reals integer, parameter :: r8 = r4 ! alias r8 to r4
Copy the tutorial DART namelist from
$DART_DIR/models/wrf/tutorial/template/input.nml.templateto$DART_DIR/models/wrf/work/input.nml.cd $DART_DIR/models/wrf cp tutorial/template/input.nml.template work/input.nml
Build the WRF-DART executables:
cd $DART_DIR/models/wrf/work ./quickbuild.sh
Many executables are built, the following executables are needed for the tutorial and will be copied to the right place by the setup.csh script in a subsequent step:
advance_time fill_inflation_restart filter obs_diag obs_seq_to_netcdf obs_sequence_tool pert_wrf_bc wrf_dart_obs_preprocess
Preparing the experiment directory.
Approximately 100Gb of space is needed to run the tutorial. Create a “work” directory someplace with a lot of free space. The rest of the instructions assume you have an environment variable called BASE_DIR that points to this directory. On Derecho it is convenient to use your scratch directory for this purpose.
shell |
command |
|---|---|
tcsh |
|
bash |
|
The WRF boundary conditions and perturbations required to make a viable ensemble are available in a 15 GB tar file. Put this file in your
$BASE_DIR. Since this is a large file, we suggest using ‘wget’ to download the file directly to your local system:cd $BASE_DIR wget http://www.image.ucar.edu/wrfdart/tutorial/wrf_dart_tutorial_23May2018_v3.tar.gz tar -xzvf wrf_dart_tutorial_23May2018_v3.tar.gz
After untarring the file you should see the following directories: icbc, output, perts, and template. The directory names (case sensitive) are important, as the scripts rely on these local paths and file names.
You will need template WRF namelists from the
$DART_DIR/models/wrf/tutorial/templatedirectory:cp $DART_DIR/models/wrf/tutorial/template/namelist.input.meso $BASE_DIR/template/. cp $DART_DIR/models/wrf/tutorial/template/namelist.wps.template $BASE_DIR/template/.
You will also need scripting to run a WRF/DART experiment. Copy the contents of
$DART_DIR/models/wrf/shell_scriptsto the$BASE_DIR/scriptsdirectory.mkdir $BASE_DIR/scripts cp -R $DART_DIR/models/wrf/shell_scripts/* $BASE_DIR/scripts
Build or locate the WRF, WPS and WRFDA executables
Instruction for donwloading the WRF package is located here. The WRF package consists of 3 parts: the WRF atmospheric model WRF(ARW), the WRF Preprocessing System (WPS) and WRF Data Assimilation System (WRFDA).
Importantly, DART is used to perform the ensemble DA for this tutorial, however,
the WRFDA package is required to generate a set of perturbed initial ensemble member
files and also to generate perturbed boundary condition files. Since the
tutorial provides a perturbation bank for a specific case, it is not
required to actually run da_wrfvar.exe but it needs to be in the
WRF_RUN directory for the tutorial.
WRF and WRFDA should be built with the “dmpar” option, while WPS can be built “serial”ly. See the WRF documentation for more information about building these packages.
Warning
For consistency and to avoid errors, you should build WRF, WPS, WRFDA, and DART with the same compiler you use for NetCDF. Likewise MPI should use the same compiler. You will need the location of the WRF and WRFDA builds to customize the params.csh script in the next step. If using gfortran to compile WRF on Derecho we recommend using option 34 (gnu dmpar) to configure WRF, option 1 (gnu serial) to configure WPS, and option 34 (gnu dmpar) to configure WRFDA. You will need the location of the WRF, WPS,and WRFDA builds to customize the params.csh script in the next step.
Using the gfortan compiler on Derecho required custom flag settings to successfully compile the WRF, WPS and WRFDA executables. For more information please see NCAR/DART github issue 627.
Configure $BASE_DIR/scripts/param.csh with proper paths, info, etc.
This is a script that sets variables which will be read by other WRF-DART scripts. There are some specific parameters for either the Derecho supercomputing system using the PBS queueing system or the (decommissioned) Yellowstone system which used the LSF queueing system. If you are not using Derecho, you may still want to use this script to set your queueing-system specific parameters.
Important
All variables that are marked
'set this appropriately #%%%#'need to be set. This list is intended to provide some guidance on what needs to be set, but it is not an exhaustive list.
Script variable |
Description |
|---|---|
module load nco |
The nco package. |
module load ncl/6.6.2 |
The ncl package. |
BASE_DIR |
The directory containing icbc, output, perts, etc. |
DART_DIR |
The DART directory. |
WRF_DM_SRC_DIR |
The directory of the WRF dmpar installation. |
WPS_SRC_DIR |
The directory of the WPS installation. |
VAR_SRC_DIR |
The directory of the WRFDA installation. |
GEO_FILES_DIR |
The root directory of the WPS_GEOG files. NOTE: on Derecho these are available in the /glade/u/home/wrfhelp/WPS_GEOG directory |
GRIB_DATA_DIR |
The root directory of the GRIB data input into ungrib.exe. For this tutorial the grib files are included, so use ${ICBC_DIR}/grib_data |
GRIB_SRC |
The type of GRIB data (e.g. <Vtable.TYPE>) to use with ungrib.exe to copy the appropriate Vtable file. For the tutorial, the value should be ‘GFS’. |
COMPUTER_CHARGE_ACCOUNT |
The project account for supercomputing charges. See your supercomputing project administrator for more information. |
The e-mail address used by the queueing system to send job summary information. This is optional. |
Run the setup.csh script to create the proper directory structure and move executables to proper locations.
cd $BASE_DIR/scripts
./setup.csh param.csh
So far, your $BASE_DIR should contain the following directories:
icbc
obs_diag
obsproc
output
perts
post
rundir
scripts
template
Your $BASE_DIR/rundir directory should contain the following:
executables:
pert_wrf_bc(no helper page),
directories:
WRFIN(empty)WRFOUT(empty)WRF_RUN(wrf executables and support files)
scripts:
add_bank_perts.ncl
new_advance_model.csh
support data:
sampling_error_correction_table.nc
Check to make sure your $BASE_DIR/rundir/WRF_RUN directory contains:
da_wrfvar.exe
wrf.exe
real.exe
be.dat
contents of your WRF build run/ directory (support data files for WRF)
Note
Be aware that the setup.csh script is designed to remove
$BASE_DIR/rundir/WRF_RUN/namelist.input. Subsequent scripting will
modify $BASE_DIR/template/namlist.input.meso to create the
namelist.input for the experiment.
For this tutorial, we are providing you with a specified WRF domain. To make your own, you would need to define your own wps namelist and use WPS to make your own geogrid files. See the WRF site for help with building and running those tools as needed. You would also need to get the appropriate grib files to generate initial and boundary condition files for the full period you plan to cycle. In this tutorial we have provided you with geogrid files, a small set of grib files, and a namelist to generate series of analyses for several days covering a North American region.
Let’s now look inside the $BASE_DIR/scripts directory. You should
find the following scripts:
Script name |
Description |
|---|---|
add_bank_perts.ncl |
Adds perturbations to each member. |
assim_advance.csh |
Advances 1 WRF ensemble member to the next analysis time. |
assimilate.csh |
Runs filter … i.e. the assimilation. |
diagnostics_obs.csh |
Computes observation-space diagnostics and the model-space mean analysis increment. |
driver.csh |
Primary script for running the cycled analysis system. |
first_advance.csh |
Advances 1 WRF ensemble member (on the first time). |
gen_pert_bank.csh |
Saves the perturbations generated by WRFDA CV3. |
gen_retro_icbc.csh |
Generates the wrfinput and wrfbdy files. |
init_ensemble_var.csh |
Creates the perturbed initial conditions from the WRF-VAR system. |
mean_increment.ncl |
Computes the mean state-space increment, which can be used for plotting. |
new_advance_model.csh |
advances the WRF model after running DART in a cycling context. |
param.csh |
Contains most of the key settings to run the WRF-DART system. |
prep_ic.csh |
Prepares the initial conditions for a single ensemble member. |
real.csh |
Runs the WRF real.exe program. |
setup.csh |
Creates the proper directory structure and place executables/scripts in proper locations. |
You will need to edit the following scripts to provide the paths to
where you are running the experiment, to connect up files, and to set
desired dates. Search for the string 'set this appropriately #%%%#'
for locations that you need to edit.
cd $BASE_DIR/scripts
grep -r 'set this appropriately #%%%#' .
Other than param.csh, which was covered above, make the following changes:
File name |
Variable / value |
Change description |
|---|---|---|
driver.csh |
datefnl = 2017042712 |
Change to the final target date; here the final date is already set correctly for this tutorial. |
gen_retro_icbc.csh |
datefnl = 2017042712 |
Set to the final target date of the tutorial. However, it is possible (not necessary) to create WRF initial/boundary conditions to 2017043000. This is the latest date that files are included in the tutorial. |
gen_retro_icbc.csh |
paramfile = <full path to param.csh> |
The full path to param.csh. Change this on the line after the comment. While these two files are in the same directory here, in general it is helpful to have one param.csh for each experiment. |
gen_pert_bank.csh |
All changes |
As the tutorial includes a perturbation bank, you will not need to run this script for the tutorial, so you will not need to change these values. However, you should set appropriate values when you are ready to generate your own perturbation bank. |
Next, move to the $BASE_DIR/perts directory. Here you will find 100
perturbation files, called a “perturbation bank.” For your own case, you
would need to create a perturbation bank of your own. A brief
description for running the script is available inside the comments of
that file. However, again, for this tutorial, this step has already been
run for you. The $BASE_DIR/icbc directory contains a geo_em_d01.nc
file (geo information for our test domain), and grib files that will be
used to generate the initial and boundary condition files. The
$BASE_DIR/template directory should contain namelists for WRF, WPS,
and filter, along with a wrfinput file that matches what will be the
analysis domain. Finally, the $BASE_DIR/output directory contains
observations within each directory name. Template files will be placed
here once created (done below), and as we get into the cycling the
output will go in these directories.
Step 2: Initial conditions
To get an initial set of ensemble files, depending on the size of your ensemble and data available to you, you might have options to initialize the ensemble from, say, a global ensemble set of states. Here, we develop a set of flow dependent errors by starting with random perturbations and conducting a short forecast. We will use the WRFDA random CV option 3 to provide an initial set of random errors, and since this is already available in the perturbation bank developed in the setup, we can simply add these to a deterministic GFS state. Further, lateral boundary uncertainty will come from adding a random perturbation to the forecast (target) lateral boundary state, such that after the integration the lateral boundaries have random errors.
First, we need to generate a set of GFS states and boundary conditions
that will be used in the cycling. Use
$BASE_DIR/scripts/gen_retro_icbc.csh to create this set of files,
which will be added to a subdirectory corresponding to the date of the
run in the $BASE_DIR/output directory. Make sure
gen_retro_icbc.csh has the appropriate path to your param.csh
script. If the param.csh script also has the correct edits for paths
and you have the executables placed in the rundir, etc., then running
gen_retro_icbc.csh should execute a series of operations to extract
the grib data, run metgrid, and then twice execute real.exe to
generate a pair of WRF files and a boundary file for each analysis time.
cd $BASE_DIR/scripts
./gen_retro_icbc.csh
Note
Ignore any rm: No match errors, as the script attempts to
delete output files if they already exist, and they will not for the
first run.
Once the script completes, inside your $BASE_DIR/output/2017042700
directory you should see these files:
wrfbdy_d01_152057_21600_mean
wrfinput_d01_152057_0_mean
wrfinput_d01_152057_21600_mean
These filenames include the Gregorian dates for these files, which is used by the dart software for time schedules. Similar files (with different dates) should appear in all of the date directories between the datea and datef dates set in the gen_retro_icbc.csh script. All directories with later dates will also have an observation sequence file obs_seq.out that contains observations to be assimilated at that time.
Next, we will execute the script to generate an initial ensemble of states for the first analysis. For this we run the script init_ensemble_var.csh, which takes two arguments: a date string and the location of the param.csh script.
cd $BASE_DIR/scripts
./init_ensemble_var.csh 2017042700 param.csh
This script generates 50 small scripts and submits them to the batch system. It assumes a PBS batch system and the ‘qsub’ command for submitting jobs. If you have a different batch system, edit this script and look near the end. You will need to modify the lines staring with #PBS and change ‘qsub’ to the right command for your system. You might also want to modify this script to test running a single member first — just in case you have some debugging to do.
However, be warned that to successfully complete the tutorial, including running the driver.csh script in Step 5, using a smaller ensemble (e.g. < 20 members) can lead to spurious updates during the analysis step, causing the WRF simulation to fail.
When complete for the full ensemble, you should find 50 new files in the
directory output/2017042700/PRIORS with names like prior_d01.0001,
prior_d01.0002, etc… You may receive an e-mail to helpfully inform
you when each ensemble member has finished.
Step 3: Prepare observations [OPTIONAL]
Warning
The observation sequence files to run this tutorial are already provided
for you. If you want to run with the provided tutorial observations, you
can skip to Step 4 right now. If you are interested in using custom
observations for a WRF experiment other than the tutorial you should read on.
The remaining instructions provided below in Step 3 are meant as a guideline
to converting raw PREPBUFR data files into the required obs_seq format
required by DART. Be aware that there is ongoing discussion of the proper
archived data set (RDA ds090.0 or ds337.0) that should be used to obtain
the PREPBUFR data. See the discussion in bug report #634.
If you have questions please contact the DART team.
Observation processing is critical to the success of running DART and is covered in Getting Started. In brief, to add your own observations to WRF-DART you will need to understand the relationship between observation definitions and observation sequences, observation types and observation quantities, and understand how observation converters extract observations from their native formats into the DART specific format.
The observation sequence files that are provided in this tutorial come from NCEP BUFR observations from the GDAS system. These observations contain a wide array of observation types from many platforms within a single file.
If you wanted to generate your own observation sequence files from PREPBUFR for an experiment with WRF-DART, you should follow the guidance on the prepbufr page to build the bufr conversion programs, get observation files for the dates you plan to build an analysis for, and run the codes to generate an observation sequence file.
The steps listed below to generate these observation sequence files are meant as a guideline for NSF NCAR Research Data Archive data file ds090.0. Be aware not all required software has been migrated to Derecho to perform this conversion. See bug report #634 for more updated information.
To reproduce the observation sequence files in the output directories, you would do the following:
Go into your DART prep_bufr observation converter directory and install the PREPBUFR utilities as follows:
cd $DART_DIR/observations/obs_converters/NCEP/prep_bufr ./install.sh
You may need to edit the install.sh script to match your compiler and system settings.
Go to the
$DART_DIR/observations/obs_converters/NCEP/prep_bufr/work/directory and run quickbuild.sh to build the DART PREPBUFR-to-intermediate-file observation processor:cd $DART_DIR/observations/obs_converters/NCEP/prep_bufr/work ./quickbuild.sh
Download the PREPBUFR observations for your desired time. Go to the NSF NCAR Research Data Archive page for the NCEP/NSF NCAR Global Reanalysis Products. Register on the site, click on the “Data Access” tab, and follow either the instructions for external users or NSF NCAR internal users.
The downloaded .tar file will often be COS-blocked. If so, the file will appear corrupted if you attempt to untar it without converting the data. See the NSF NCAR COS-block page for more information on how to strip the COS-blocking off of your downloaded file.
Untar the data in your desired directory.
In the
$DART_DIR/observations/obs_converters/NCEP/prep_bufr/workdirectory, edit the input.nml file. This file will control what observations will be used for your experiment, so the namelist options are worth investigating a bit here. For example, you could use the following:&prep_bufr_nml obs_window = 1.0 obs_window_cw = 1.5 otype_use = 120.0, 130.0, 131.0, 132.0, 133.0, 180.0 181.0, 182.0, 220.0, 221.0, 230.0, 231.0 232.0, 233.0, 242.0, 243.0, 245.0, 246.0 252.0, 253.0, 255.0, 280.0, 281.0, 282.0 qctype_use = 0,1,2,3,15 /
This defines an observation time window of +/- 1.0 hours, while cloud motion vectors will be used over a window of +/- 1.5 hours. This will use observation types sounding temps (120), aircraft temps (130,131), dropsonde temps (132), mdcars aircraft temps, marine temp (180), land humidity (181), ship humidity (182), rawinsonde U,V (220), pibal U,V (221), Aircraft U,V (230,231,232), cloudsat winds (242,243,245), GOES water vapor (246), sat winds (252,253,255), and ship obs (280, 281, 282). Additionally, it will include observations with specified qc types only. See the prepbufr page for more available namelist controls.
Within the
$DART_DIR/observations/obs_converters/NCEP/prep_bufr/workdirectory, edit the prepbufr.csh file and change BUFR_dir, BUFR_idir, BUFR_odir, and BUFR_in to match the locations and format of the data you downloaded. A little trial and error might be necessary to get these set correctly.Copy over the executables from
../exe, and run the prepbufr.csh script for a single day at a time:cd $DART_DIR/observations/obs_converters/NCEP/prep_bufr/work cp ../exe/\*.x . ./prepbufr.csh \<year\> \<month\> \<day\>
Your PREPBUFR files have now been converted to an intermediate ASCII format. There is another observation converter to take the observations from this format and write them into the native DART format. Edit the input.nml namelist file in the DART_DIR/observations/obs_converters/NCEP/ascii_to_obs/work directory. Here is a basic example:
&ncepobs_nml year = 2017, month = 4, day = 27, tot_days = 3, max_num = 800000, select_obs = 0, ObsBase = '<path to observations>/temp_obs.', daily_file = .false., lat1 = 15.0, lat2 = 60.0, lon1 = 270.0, lon2 = 330.0 /
Choosing “select_obs = 0” will select all the observations in the ASCII file. Set “ObsBase” to the directory you output the files from during the last step. If you wish to choose specific observations from the ASCII intermediate file or control other program behavior, there are many namelist options documented on the create_real_obs page.
It is now time to build ascii_to_obs programs. Run the following:
cd $DART_DIR/observations/obs_converters/NCEP/ascii_to_obs/work ./quickbuild.sh
Run the create_real_obs program to create the DART observation sequence files:
cd $DART_DIR/observations/obs_converters/NCEP/ascii_to_obs/work ./create_real_obs
The program create_real_obs will create observation sequence files with one file for each six hour window. For a cycled experiment, the typical approach is to put a single set of observations, associated with a single analysis step, into a separate directory. For example, within the
outputdirectory, we would create directories like2017042700,2017042706,2017042712, etc. for 6-hourly cycling. Place the observation files in the appropriate directory to match the contents in the files (e.g. obs_seq2017042706) and rename as simply obs_seq.out (e.g.output/2017042706/obs_seq.out).It is helpful to also run the wrf_dart_obs_preprocess program, which can strip away observations not in the model domain, perform superobservations of dense observations, increase observation errors near the lateral boundaries, check for surface observations far from the model terrain height, and other helpful pre-processing steps. These collectively improve system performance and simplify interpreting the observation space diagnostics. There are a number of namelist options to consider, and you must provide a wrfinput file for the program to access the analysis domain information.
Step 4: Creating the first set of adaptive inflation files
In this section we describe how to create initial adaptive inflation files. These will be used by DART to control how the ensemble is inflated during the first assimilation cycle.
It is convenient to create initial inflation files before you start an experiment. The initial inflation files may be created with fill_inflation_restart, which was built by the quickbuild.sh step. A pair of inflation files is needed for each WRF domain.
Within the $BASE_DIR/rundir directory, the input.nml file has some
settings that control the behavior of fill_inflation_restart. Within
this file there is the section:
&fill_inflation_restart_nml
write_prior_inf = .true.
prior_inf_mean = 1.00
prior_inf_sd = 0.6
write_post_inf = .false.
post_inf_mean = 1.00
post_inf_sd = 0.6
input_state_files = 'wrfinput_d01'
single_file = .false.
verbose = .false.
/
These settings write a prior inflation file with a inflation mean of 1.0
and a prior inflation standard deviation of 0.6. These are reasonable
defaults to use. The input_state_files variable controls which file to
use as a template. You can either modify this namelist value to point to
one of the wrfinput_d01_XXX files under $BASE_DIR/output/<DATE>,
for any given date, or you can copy one of the files to this directory.
The actual contents of the file referenced by input_state_files do not
matter, as this is only used as a template for the
fill_inflation_restart program to write the default inflation values.
Note that the number of files specified by input_state_files must
match the number of domains specified in model_nml:num_domains, i.e.
the program needs one template for each domain. This is a
comma-separated list of strings in single ‘quotes’.
After running the program, the inflation files must then be moved to the directory expected by the driver.csh script.
Run the following commands with the dates for this particular tutorial:
cd $BASE_DIR/rundir
cp ../output/2017042700/wrfinput_d01_152057_0_mean ./wrfinput_d01
./fill_inflation_restart
mkdir ../output/2017042700/Inflation_input
mv input_priorinf_*.nc ../output/2017042700/Inflation_input/
Once these files are in the right place, the scripting should take care of renaming the output from the previous cycle as the input for the next cycle.
Step 5: Cycled analysis system
While the DART system provides executables to perform individual tasks necessary for ensemble data assimilation, for large models such as WRF that are run on a supercomputer queueing system, an additional layer of scripts is necessary to glue all of the pieces together. A set of scripts is provided with the tutorial tarball to provide you a starting point for your own WRF-DART system. You will need to edit these scripts, perhaps extensively, to run them within your particular computing environment. If you will run on NSF NCAR’s Derecho environment, fewer edits may be needed, but you should familiarize yourself with running jobs on Derecho if necessary. A single forecast/assimilation cycle of this tutorial can take up to 10 minutes on Derecho - longer if debug options are enabled or if there is a wait time during the queue submission.
In this tutorial, we have previously edited the param.csh and other scripts. Throughout the WRF-DART scripts, there are many options to adjust cycling frequency, domains, ensemble size, etc., which are available when adapting this set of scripts for your own research. To become more famililar with this set of scripts and to eventually make these scripts your own, we advise commenting out all the places the script submits jobs while debugging, placing an ‘exit’ in the script at each job submission step. This way you will be able to understand how all of the pieces work together.
However, for this tutorial, we will only show you how the major components work. The next step in our process is the main driver.csh script, which expects a starting date (YYYYMMDDHH) and the full path of the resource file as command line arguments. In this example (which uses csh/tcsh syntax), we are also capturing the run-time output into a file named run.out and the entire command will be running in the background:
cd $BASE_DIR/scripts
./driver.csh 2017042706 param.csh >& run.out &
driver.csh will - check that the input files are present (wrfinput
files, wrfbdy, observation sequence, and DART restart files), - create a
job script to run filter in $BASE_DIR/rundir, - monitor that
expected output from filter is created, - submit jobs to advance the
ensemble to the next analysis time, - (simultaneously with the ensemble
advance) compute assimilation diagnostics - archive and clean up - and
continue to cycle until the final analysis time has been reached.
Step 6: Diagnosing the assimilation results
Once you have successfully completed steps 1-5, it is important to check the quality of the assimilation. In order to do this, DART provides analysis system diagnostics in both state and observation space.
As a preliminary check, confirm that the analysis system actually updated
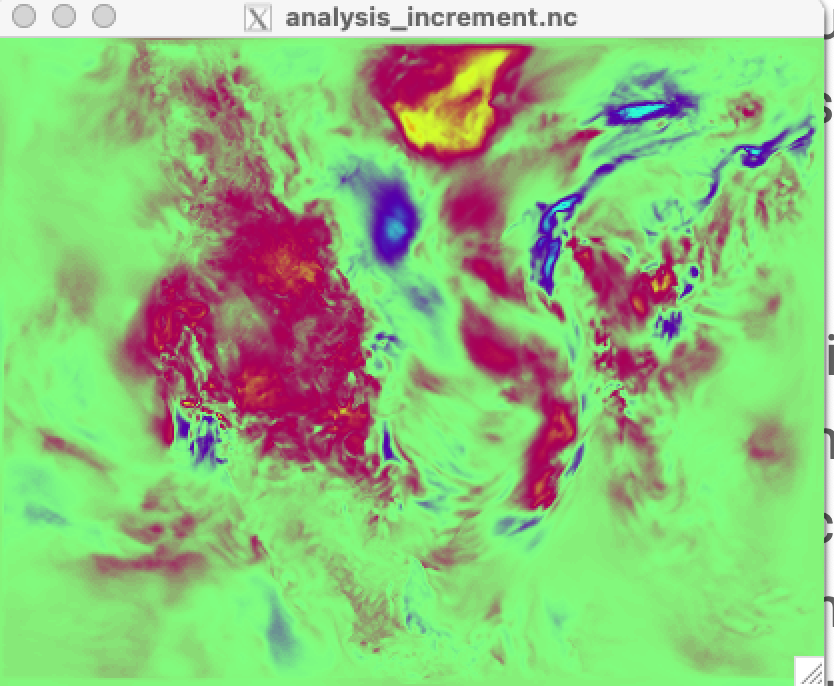
the WRF state. Locate the file in the $BASE_DIR/output/* directory called
analysis_increment.nc which is the difference of the ensemble mean state
between the background (prior) and the analysis (posterior) after running
filter. Use a tool, such as ncview, to look at this file as follows:
cd $BASE_DIR/output/datefnl
module load ncview
ncview analysis_increment.nc
The analysis_increment.nc file includes the following atmospheric variables:
MU, PH, PSFC, QRAIN, QCLOUD, QGRAUP, QICE, QNICE, QSNOW, QVAPOR, T and T2.
The example figure below shows the increments for temperature (T) only. You can
use ncview to advance through all 11 atmospheric pressure levels. You should
see spatial patterns that look something like the meteorology of the day.
For more information on how the increments were calculated, we recommend (but do not require to complete the tutorial) that you review the Diagnostics Section of the DART Documentation. There are seven sections within the diagnostics section including 1) Checking your initial assimilation, 2) Computing filter increments and so on. Be sure to advance through all the sections.
The existence of increments proves the model state was adjusted, however, this says nothing about the quality of the assimilation. For example, how many of the observations were assimilated? Does the posterior state better represent the observed conditions of the atmosphere? These questions can be addressed with the tools described in the remainder of this section. All of the diagnostic files (obs_epoch*.nc and obs_diag_output.nc) have already been generated from the tutorial. (driver.csh* executes **diagnostics_obs.csh). Therefore you are ready to start the next sections.
Visualizing the observation locations and acceptance rate
An important assimilation diagnostic is whether observations were accepted or rejected. Observations can be rejected for many reasons, but the two most common rejection modes in DART are: 1) violation of the outlier threshold, meaning the observations were too far away from the prior model estimate of the observation or 2) forward operator failure, meaning the calculation to generate the expected observation failed. A full list of rejection criteria are provided here. Regardless of the reason for the failure, a successful simulation assimilates the vast majority of observations. The tools below provide methods to visualize the spatial patterns, statistics and failure mode for all observations.
The observation diagnostics use the obs_epoch*.nc file as input. This file is automatically generated by the obs_diagnostic.csh script within Step 5 of this tutorial.
The obs_epoch*.nc file is located in the output directory of each time step. In some cases there could be multiple obs_epoch*.nc files, but in general, the user should use the obs_epoch file appended with the largest numeric value as it contains the most complete set of observations. The diagnostic scripts used here are included within the DART package, and require a license of Matlab to run. The commands shown below to run the diagnostics use NSF NCAR’s Derecho, but a user could also run on their local machine.
First explore the obs_epoch*.nc file and identify the variety of observations included in the assimilation including aircraft, surface, satelllite and radiosonde types.
ncdump -h $BASEDIR/output/datefnl/obs_epoch_029.nc
..
..
RADIOSONDE_U_WIND_COMPONENT
RADIOSONDE_V_WIND_COMPONENT
RADIOSONDE_TEMPERATURE
RADIOSONDE_SPECIFIC_HUMIDITY
ACARS_U_WIND_COMPONENT
ACARS_V_WIND_COMPONENT
ACARS_TEMPERATURE
MARINE_SFC_U_WIND_COMPONENT
MARINE_SFC_V_WIND_COMPONENT
MARINE_SFC_TEMPERATURE
MARINE_SFC_SPECIFIC_HUMIDITY
LAND_SFC_U_WIND_COMPONENT
LAND_SFC_V_WIND_COMPONENT
LAND_SFC_TEMPERATURE
LAND_SFC_SPECIFIC_HUMIDITY
SAT_U_WIND_COMPONENT
SAT_V_WIND_COMPONENT
RADIOSONDE_SURFACE_ALTIMETER
MARINE_SFC_ALTIMETER
LAND_SFC_ALTIMETER
METAR_ALTIMETER
METAR_U_10_METER_WIND
METAR_V_10_METER_WIND
METAR_TEMPERATURE_2_METER
METAR_SPECIFIC_HUMIDITY_2_METER
METAR_DEWPOINT_2_METER
RADIOSONDE_DEWPOINT
LAND_SFC_DEWPOINT
RADIOSONDE_RELATIVE_HUMIDITY
LAND_SFC_RELATIVE_HUMIDITY
..
..
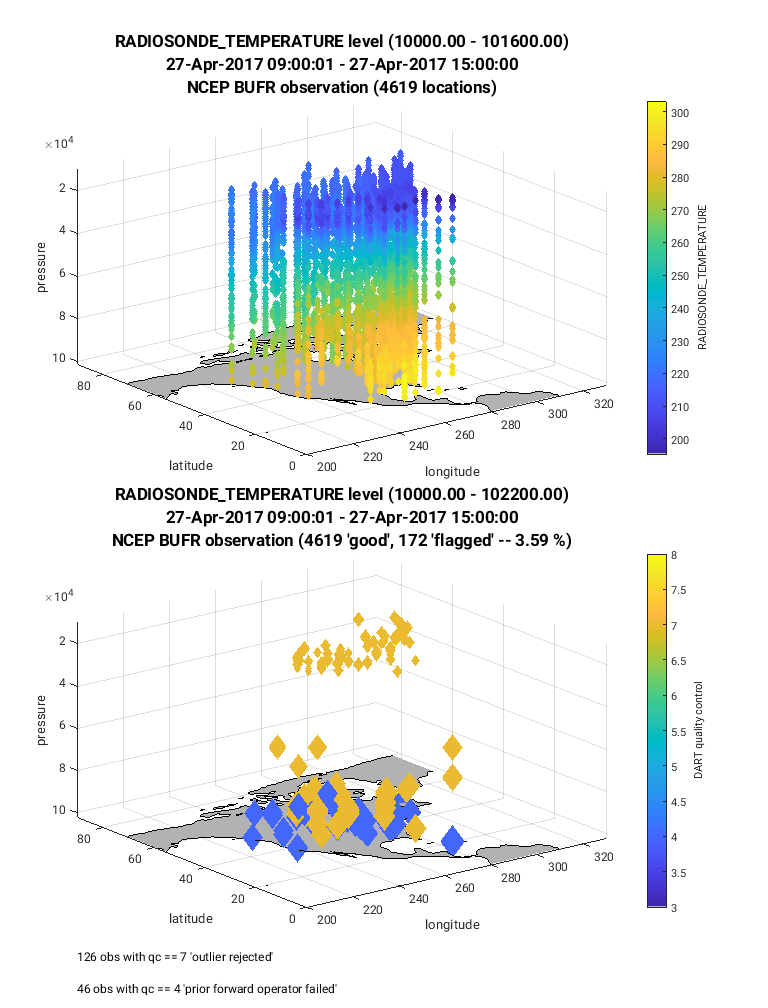
The example below uses the plot_obs_netcdf.m script to visulaize
the observation type: RADIOSONDE_TEMPERATURE which includes both horizontal
and vertical coverage across North America. We recommend to view the script’s
contents with a text editor, paying special attention to the beginning of the file
which is notated with a variety of examples. Then to run the example do the
following:
cd $DARTROOT/diagnostics/matlab
module load matlab
matlab -nodesktop
Within Matlab declare the following variables, then run the script
plot_obs_netcdf.m as follows below being sure to modify the
fname variable for your specific case.
>> fname = '$BASEDIR/output/2017042712/obs_epoch_029.nc';
>> ObsTypeString = 'RADIOSONDE_TEMPERATURE';
>> region = [200 330 0 90 -Inf Inf];
>> CopyString = 'NCEP BUFR observation';
>> QCString = 'DART quality control';
>> maxgoodQC = 2;
>> verbose = 1; % anything > 0 == 'true'
>> twoup = 1; % anything > 0 == 'true'
>> plotdat = plot_obs_netcdf(fname, ObsTypeString, region, CopyString, QCString, maxgoodQC, verbose, twoup);
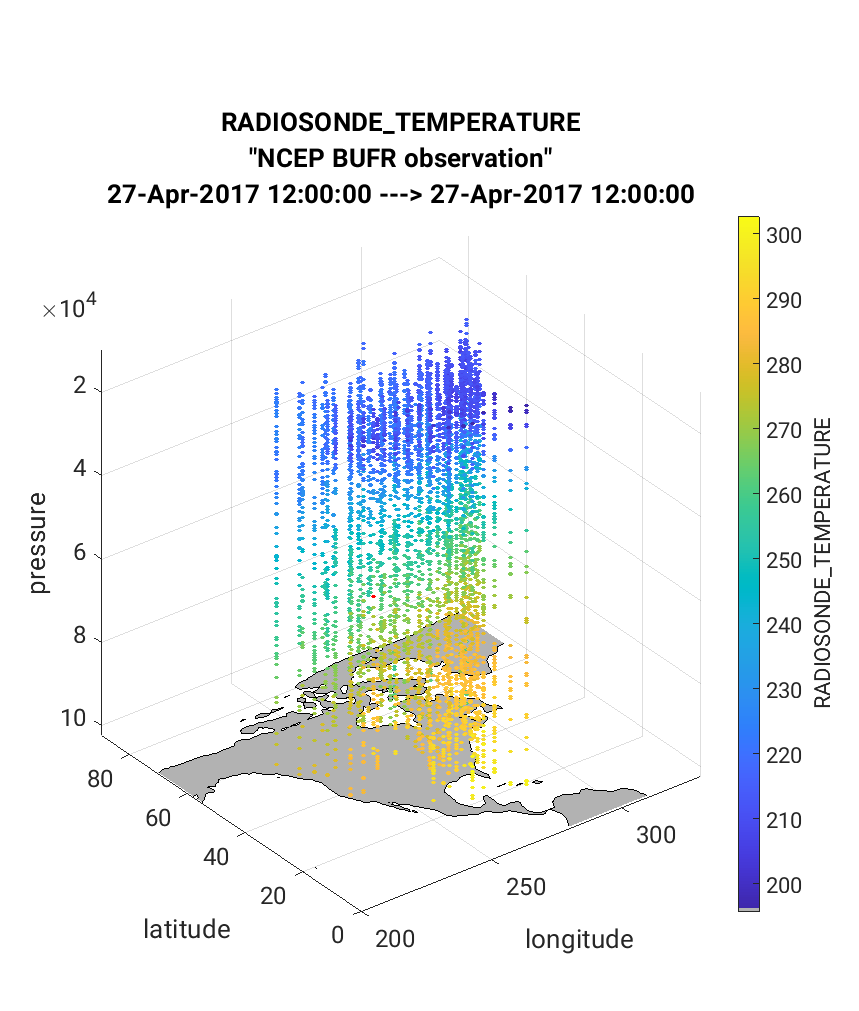
Below is an example of the figure produced by plot_obs_netcdf.m.
Note that the top panel includes both the 3-D location of all possible
RADIOSONDE_TEMPERATURE observations, which are color-coded based upon
the temperature value. The bottom panel, on the other hand, provides only
the location of the observations that were rejected by the assimilation.
The color code indicates the reason for the rejection based on the
DART quality control (QC).
In this example observations were rejected based on violation of the
outlier threshold (QC = 7), and forward operator failure (QC = 4).
Text is included within the figures that give more details regarding the
rejected observations (bottom left of figure), and percentage of observations
that were rejected (flagged, located within title of figure).
Tip
The user can manually adjust the appearance of the data by accessing the ‘Rotate 3D’ option either by clicking on the top of the figure or through the menu bar as Tools > Rotate 3D. Use your cursor to rotate the map to the desired orientation.
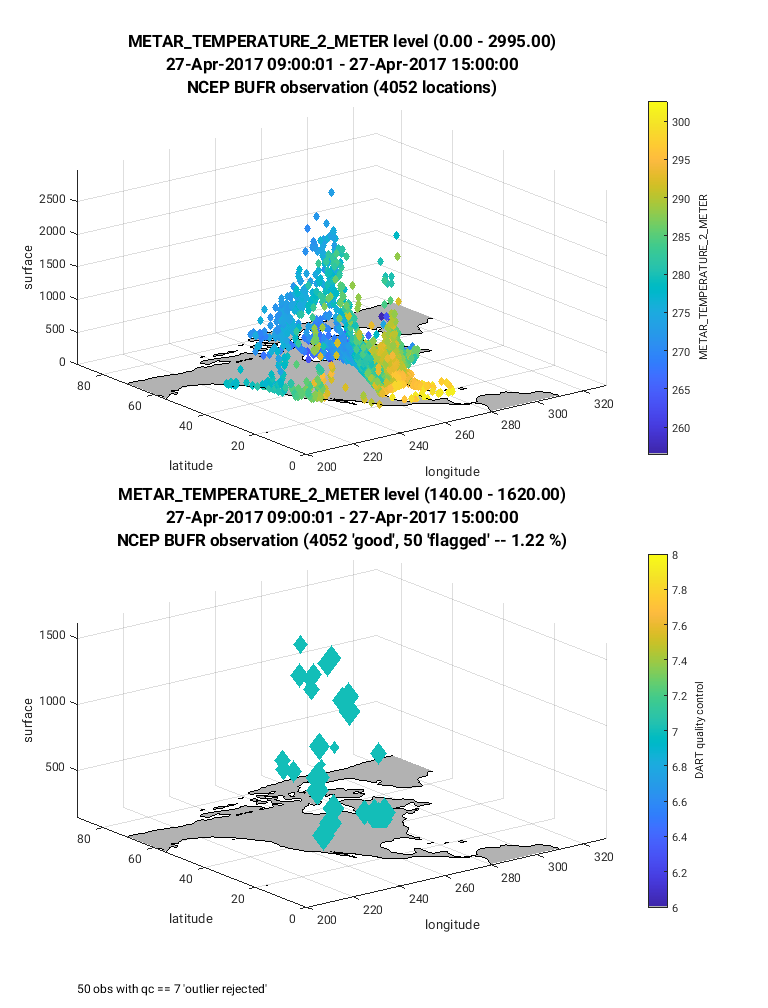
For the next figure (below) the same steps are taken as described
above, however, the observation type (ObsTypeString) is set to
METAR_TEMPERATURE_2_METER. Notice in this case the observations
are limited to near the land surface. This is because the vertical location
of this observation type was defined to be at the land surface
(VERTISSURFACE), as opposed to the RADIOSONDE_TEMPERATURE observation
in which the vertical location was defined as pressure (VERTISPRESSURE). The
vertical coordinate system is defined in the obs_seq.out file and
documented here.
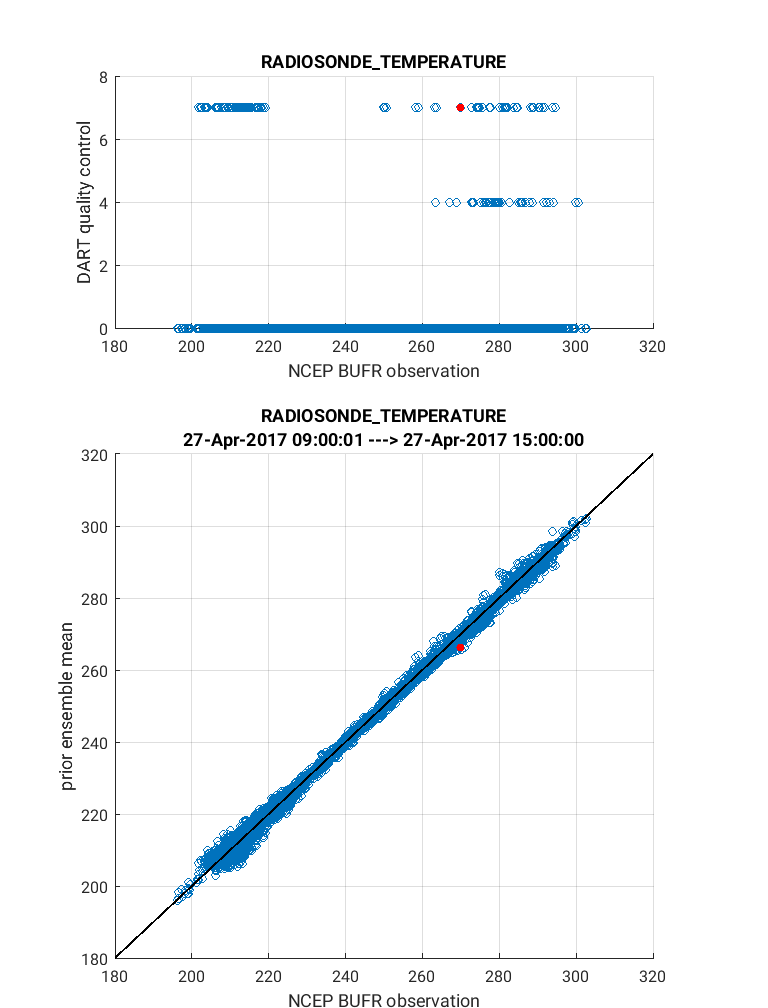
Next we will demonstrate the use of the link_obs.m script which
provides visual tools to explore how the observations impacted the
assimilation. The script generates 3 different figures which includes
a unique linking feature that allows the user to identify the features
of a specific observation including physical location, QC value, and
prior/posterior estimated values. In the example below the ‘linked’
observation appears ‘red’ in all figures. To execute link_obs.m do the
following within Matlab being sure to modify fname for your case:
>> clear all
>> close all
>> fname = '$BASEDIR/output/2017042712/obs_epoch_029.nc';
>> ObsTypeString = 'RADIOSONDE_TEMPERATURE';
>> region = [200 330 0 90 -Inf Inf];
>> ObsCopyString = 'NCEP BUFR observation';
>> CopyString = 'prior ensemble mean';
>> QCString = 'DART quality control';
>> global obsmat;
>> link_obs(fname, ObsTypeString, ObsCopyString, CopyString, QCString, region)
Tip
To access the linking feature, click near the top of the figure such that a list of icons appear. Next click on the ‘brush data’ icon then click on a data point you wish to link. It will appear red. Alternatively you can access the brush tool through the menu bar (Tools > Brush).
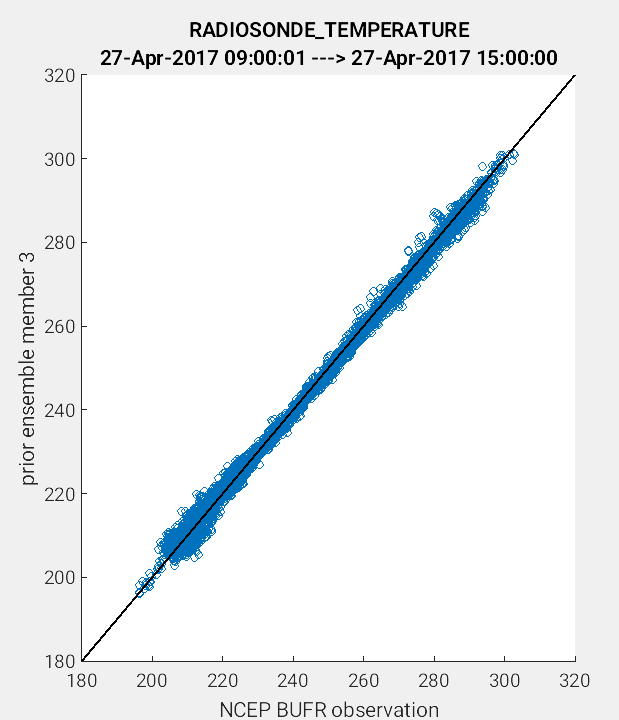
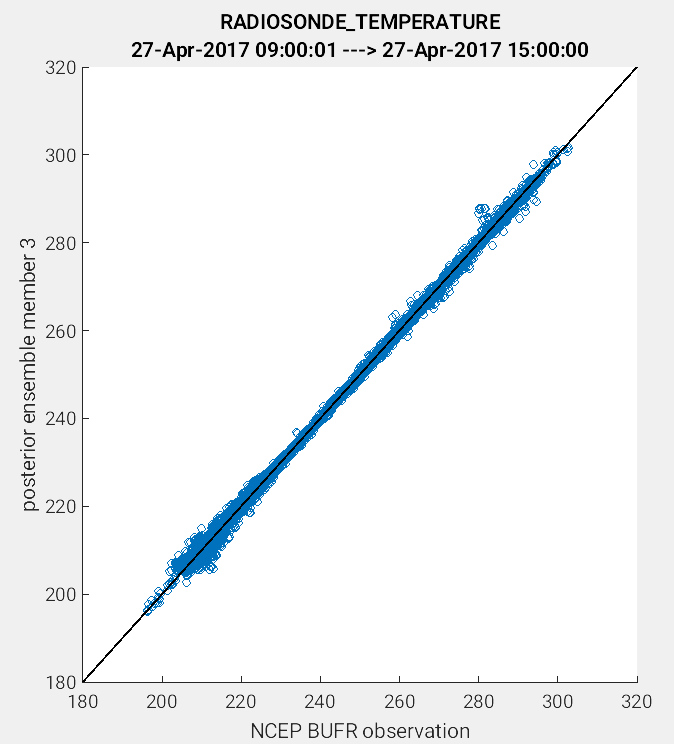
Another useful application of the link_obs.m script is to visually identify
the improvement of the model estimate of the observation through the 1:1 plot.
One way to do this is to compare the prior and posterior model estimate of the
either the ensemble mean or a single ensemble member. In the example figures below,
a 1:1 plot was generated for the prior and posterior values for ensemble member 3.
(Left Figure: CopyString = 'prior ensemble member 3' and Right Figure:
CopyString = posterior ensemble member 3'). Note how the prior member
estimate (left figure) compares less favorably to the observations as compared
to the posterior member estimate (right figure). The improved alignment
(blue circles closer to 1:1 line) between the posterior estimate and the observations
indicates that the DART filter update provided an improved representation of the
observed atmospheric state.
So far the example figures have provided primarily qualitative estimates of the assimilation performance. The next step demonstrates how to apply more quantitative measures to assess assimilation skill.
Quantification of model-observation mismatch and ensemble spread
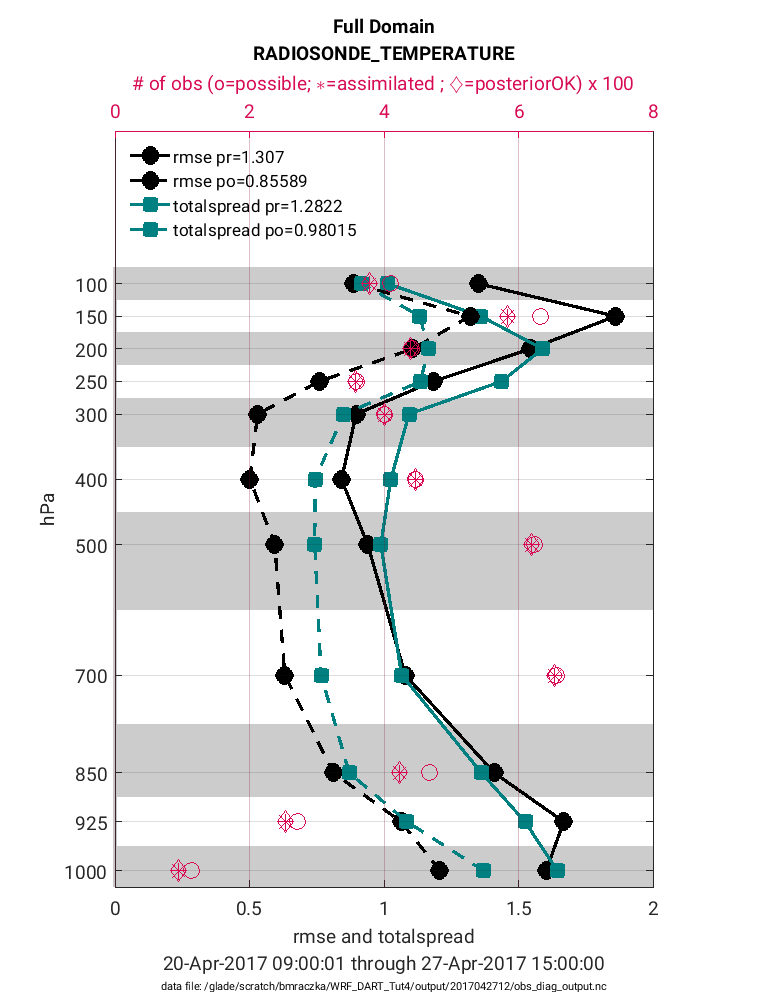
The plot_rmse_xxx_profile.nc script is one of the best tools to evaluate
assimilation performance across a 3-D domain such as the atmosphere.
It uses the obs_diag_output.nc file as an input to generate RMSE,
observation acceptance and other statistics. Here we choose the ensemble
‘total spread’ statistic to plot alongside RMSE, however, you can choose
other statistics including ‘bias’, ‘ens_mean’ and ‘spread’. For a full
list of statistics perform the command ncdump -v CopyMetaData obs_diag_output.nc.
>> fname ='$BASEDIR/output/2017042712/obs_diag_output.nc';
>> copy = 'totalspread';
>> obsname = 'RADIOSONDE_TEMPERATURE';
>> plotdat = plot_rmse_xxx_profile(fname,copy,'obsname',obsname)
Note in the figure above that the prior RMSE and total spread values (solid black and teal lines) are significantly greater than the posterior values (dashed black and teal lines). This is exactly the behavior we would expect (desire) because the decreased RMSE indicates the posterior model state has an improved representation of the atmosphere. It is common for the introduction of observations to also reduce the ‘total spread’ because the prior ensemble spread will compress to better match the observations. In general, it is preferable for the magnitude of the total spread to be similar to the RMSE. If there are strong departures between the total spread and RMSE this suggests the adaptive inflation settings may need to be adjusted to avoid filter divergence. Note that these statistics are given for each pressure level (1-11) within the WRF model. Accompanying each level is also the total possible (pink circle) and total assimilated (pink asterisk) observations. Note that for each level the percentage of assimilated observations is quite high (>90%). This high acceptance percentage is typical of a high-quality assimilation and consistent with the strong reduction in RMSE.
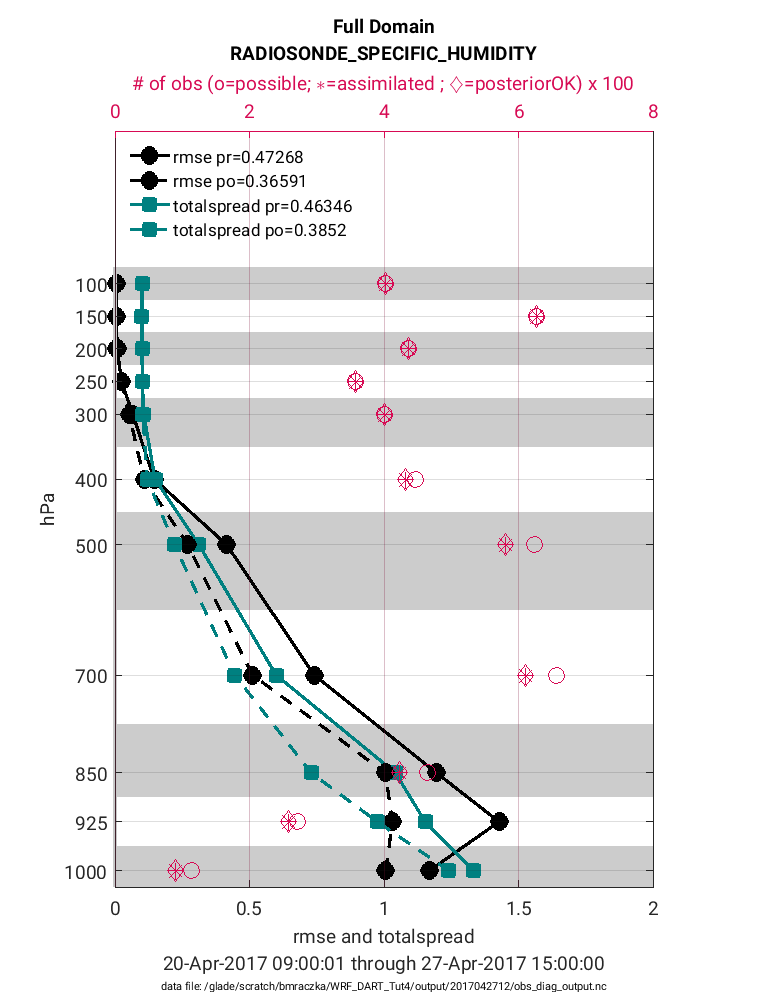
The same plot as above is given below except for the observation type:
RADIOSONE_SPECIFIC_HUMIDITY.
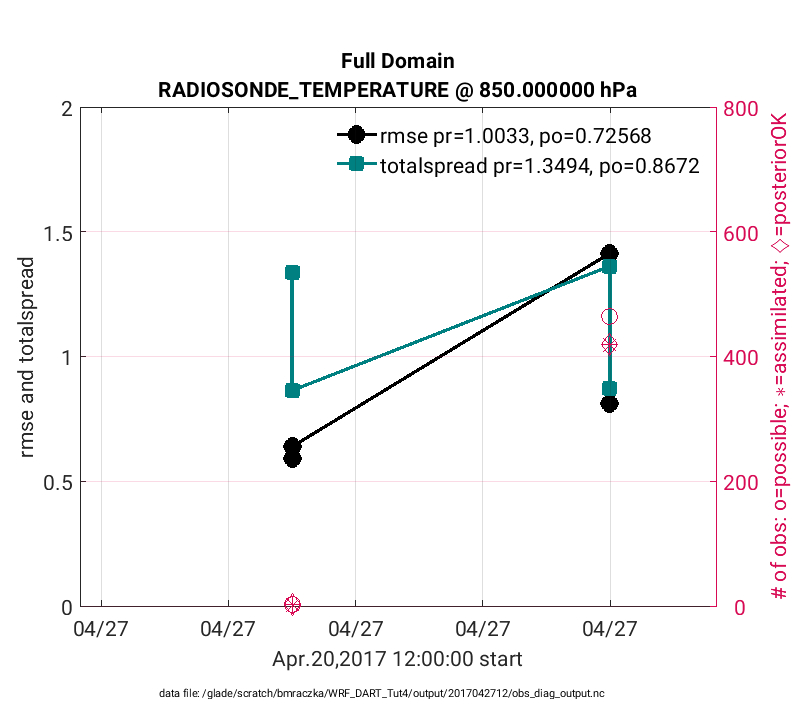
Although the plot_rmse_xxx_profile.m script is valuable for visualizing vertical profiles of assimilation statistics, it doesn’t capture the temporal evolution. Temporal evolving statistics are valuable because the skill of an assimilation often begins poorly because of biases between the model and observations, which should improve with time. Also the quality of the assimilation may change because of changes in the quality of the observations. In these cases the plot_rmse_xxx_evolution.m script is used to illustrate temporal changes in assimilation skill. To generate the figures below the following matlab commands were used:
>> fname = '$BASEDIR/output/2017042712/obs_diag_output.nc';
>> copy = 'totalspread';
>> obsname = 'RADIOSONDE_TEMPERATURE';
>> plotdat = plot_rmse_xxx_evolution(fname,copy,'obsname',obsname,'level',3);
Note
The figures below only evaluate two different assimilation
cycles (hour 6 and hour 12 on 4/27/17), thus it is difficult to evaluate the
temporal progression of the assimilation statistics. This is given purely as an
example. Real world assimilations generally span for months and years thus
evaluating temporal evolution of statistics is more straightforward. The x-axis was
also manually adjusted in the figure below. To do this
plot_rmse_xxx_evolution.m was edited such that the bincenters were replaced
with datenum values when defining axlims as:
axlims = [datenum(2017,4,27,2,0,0) datenum(2017,4,27,14,0,0) plotdat.Yrange];
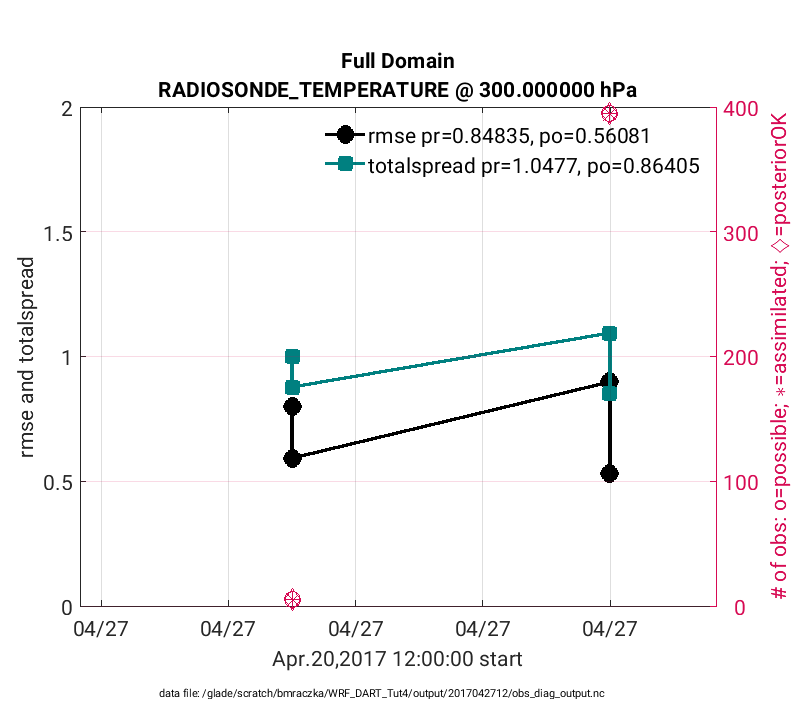
The above figure is evaluated at model level 850hPa (‘level’,3), whereas the figure below is generated in the same way except is evaluated at 300 hPa (‘level’,7) using: plotdat = plot_rmse_xxx_evolution(fname,copy,’obsname’,obsname,’level’,7)
Important
The example diagnostics provided here are only a subset of the diagnostics available in the DART package. Please see the web-based diagnostic documentation. or DART LAB and DART Tutorial for more details.
Generating the obs_diag_output.nc and obs_epoch*.nc files manually [OPTIONAL]
This step is optional because the WRF-DART Tutorial automatically generates the diagnostic files (obs_diag_output.nc and obs_epoch_*.nc). However, these files were generated with pre-set options (e.g. spatial domain, temporal bin size etc.) that you may wish to modify. Also, it is uncommon to generate these diagnostics files automatically for a new assimilation application. Therefore this section describes the steps to generate the diagnostic files directly from the DART scripts by using the WRF Tutorial as an example.
Generating the obs_epoch*.nc file
cd $DARTROOT/models/wrf/work
Generate a list of all the obs_seq.final files created by the assimilation step (filter step). This command creates a text list file.
ls /glade/scratch/bmraczka/WRF_DART_Tut4/output/2017*/obs_seq.final > obs_seq_tutorial.txt
The DART exectuable obs_seq_to_netcdf is used to generate the obs_epoch
type files. Modify the obs_seq_to_netcdf and schedule namelist settings
(using a text editor like vi) with the input.nml file to specify the spatial domain
and temporal binning. The values below are intended to include the entire time
period of the assimilation.
&obs_seq_to_netcdf_nml
obs_sequence_name = ''
obs_sequence_list = 'obs_seq_tutorial.txt',
lonlim1 = 0.0
lonlim2 = 360.0
latlim1 = -90.0
latlim2 = 90.0
verbose = .false.
/
&schedule_nml
calendar = 'Gregorian',
first_bin_start = 1601, 1, 1, 0, 0, 0,
first_bin_end = 2999, 1, 1, 0, 0, 0,
last_bin_end = 2999, 1, 1, 0, 0, 0,
bin_interval_days = 0,
bin_interval_seconds = 21600,
max_num_bins = 1000,
print_table = .true
/
Finally, run the exectuable:
./obs_seq_to_netcdf
Generating the obs_diag_output.nc file
cd $DARTROOT/models/wrf/work
The DART exectuable obs_diag is used to generate the obs_diag_output
files. Modify the obs_diag namelist settings
(using a text editor like vi) with the input.nml file to specify the spatial domain
and temporal binning. Follow the same steps to generate the obs_seq_tutorial.txt
file as described in the previous section.
&obs_diag_nml
obs_sequence_name = '',
obs_sequence_list = 'obs_seq_tutorial.txt',
first_bin_center = 2017, 4, 27, 0, 0, 0 ,
last_bin_center = 2017, 4, 27, 12, 0, 0 ,
bin_separation = 0, 0, 0, 6, 0, 0 ,
bin_width = 0, 0, 0, 6, 0, 0 ,
time_to_skip = 0, 0, 0, 0, 0, 0 ,
max_num_bins = 1000,
Nregions = 1,
lonlim1 = 0.0,
lonlim2 = 360.0,
latlim1 = 10.0,
latlim2 = 65.0,
reg_names = 'Full Domain',
print_mismatched_locs = .false.,
verbose = .true.
/
Finally, run the exectuable:
./obs_diag
If you encounter difficulties setting up, running, or evaluating the system performance, please consider using the GitHub Issue facility or feel free to contact us at dart(at)ucar(dot)edu.
Additional materials from previous in-person tutorials
Introduction - DART Lab materials
WRF-DART basic building blocks -slides (some material is outdated)
Computing environment support -slides
WRF-DART application examples -slides (some material is outdated)
Observation processing -slides
DART diagnostics - observation diagnostics
More Resources
Preparing MATLAB to use with DART.