program obs_seq_verify
Overview
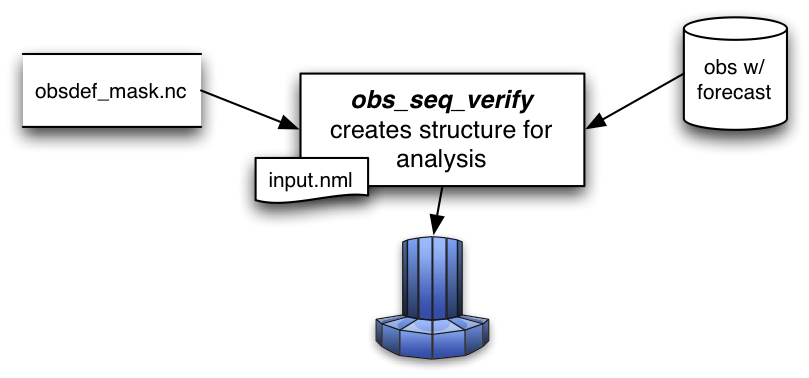
obs_seq_verify reorders the observations from a forecast run of DART into a structure that is amenable for the
evaluation of the forecast. The big picture is that the verification locations and times identified in the
obsdef_mask.nc and the observations from the forecast run (whose files must have an extension as in the
following: obs_seq.forecast.YYYYMMDDHH) are put into a netCDF variable that looks like this:obs_seq_verify can read in a series of observation sequence files - each of the files must contain the
entire forecast from a single analysis time. The extension of each filename is required to reflect the
analysis time. Use program obs_sequence_tool to concatenate
multiple files into a single observation sequence file if necessary. Only the individual ensemble members forecast
values are used - the ensemble mean and spread (as individual copies) are completely ignored. The individual “prior
ensemble member NNNN” copies are used. As a special case, the “prior ensemble mean” copy is used if and only if
there are no individual ensemble members present (i.e. input.nml &filter_nml:num_output_obs_members == 0).Dimension |
Explanation |
|---|---|
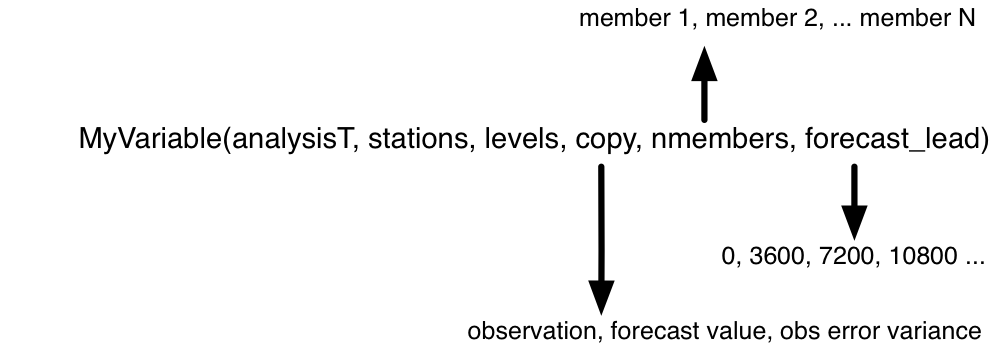
analysisT |
This is the netCDF UNLIMITED dimension, so it is easy to ‘grow’ this dimension. This corresponds to the number of forecasts one would like to compare. |
stations |
The unique horizontal locations in the verification network. |
levels |
The vertical level at each location. Observations with a pressure vertical coordinate are selected based on their proximity to the mandatory levels as defined in program obs_seq_coverage. Surface observations or observations with undefined vertical coordinates are simply put into level 1. |
copy |
This dimension designates the quantity of interest; the observation, the forecast value, or the observation error variance. These quantities are the ones required to calculate the evaluation statistics. |
nmembers |
Each ensemble member contributes a forecast value. |
forecast_lead |
This dimension relates to the amount of time between the start of the forecast and the verification. |
The USAGE section has more on the actual use of obs_seq_verify.
Namelist
This namelist is read from the file input.nml. Namelists start with an ampersand ‘&’ and terminate with a slash ‘/’.
Character strings that contain a ‘/’ must be enclosed in quotes to prevent them from prematurely terminating the
namelist.
&obs_seq_verify_nml
obs_sequences = ''
obs_sequence_list = ''
station_template = 'obsdef_mask.nc'
netcdf_out = 'forecast.nc'
obtype_string = 'RADIOSONDE_TEMPERATURE'
print_every = 10000
verbose = .true.
debug = .false.
/
You can specify either obs_sequences or obs_sequence_list – not both. One of them has to be an empty
string … i.e. ‘ ‘.
Item |
Type |
Description |
|---|---|---|
obs_sequences |
character(len=256), dimension(500) |
Names of the observation sequence files - each of which
MUST have an extension that defines the start of the
forecast (the analysis time). The observation sequence
filenames must be something like
|
obs_sequence_list |
character(len=256) |
Name of an ascii text file which contains a list of one
or more observation sequence files, one per line. The
observation sequence filenames MUST have an extension
that defines the start of the forecast (the analysis
time). The observation sequence filenames must be
something like |
station_template |
character(len=256) |
The name of the netCDF file created by program obs_seq_coverage that contains the verification network description. |
netcdf_out |
character(len=256) |
The base portion of the filename of the file that will
contain the forecast quantities. Since each observation
type of interest is processed with a separate run of
|
calendar |
character(len=129) |
The type of the calendar used to interpret the dates. |
obtype_string |
character(len=32) |
The observation type string that will be verified. The character string must match one of the standard DART observation types. This will be the name of the variable in the netCDF file, and will also be used to make a unique netCDF file name. |
print_every |
integer |
Print run-time information for every |
verbose |
logical |
Print extra run-time information. |
debug |
logical |
Print a frightening amount of run-time information. |
Other modules used
assimilation_code/location/threed_sphere/location_mod.f90
assimilation_code/modules/assimilation/assim_model_mod.f90
models/your_model/model_mod.f90
assimilation_code/modules/observations/obs_kind_mod.f90
assimilation_code/modules/observations/obs_sequence_mod.f90
assimilation_code/modules/utilities/null_mpi_utilities_mod.f90
assimilation_code/modules/utilities/types_mod.f90
assimilation_code/modules/utilities/random_seq_mod.f90
assimilation_code/modules/utilities/time_manager_mod.f90
assimilation_code/modules/utilities/utilities_mod.f90
observations/forward_operators/obs_def_mod.f90
Files
input.nmlis used for obs_seq_verify_nmlA netCDF file containing the metadata for the verification network. This file is created by program obs_seq_coverage to define the desired times and locations for the verification. (
obsdef_mask.ncis the default name)One or more observation sequence files from
filterrun in forecast mode - meaning all the observations were flagged as evaluate_only. It is required/presumed that all the ensemble members are output to the observation sequence file (see num_output_obs_members). Each observation sequence file contains all the forecasts from a single analysis time and the filename extension must reflect the analysis time used to start the forecast. (obs_seq.forecast.YYYYMMDDHHis the default name)Every execution of
obs_seq_verifyresults in one netCDF file that contains the observation being verified. Ifobtype_string = 'METAR_U_10_METER_WIND', andnetcdf_out = 'forecast.nc'; the resulting filename will beMETAR_U_10_METER_WIND_forecast.nc.
Usage
obs_seq_verify is built in …/DART/models/your_model/work, in the same way as the other DART components.obs_seq_verify sorts an unstructured, unordered set of
observations into a predetermined configuration.Example: a single 48-hour forecast that is evaluated every 6 hours
obsdef_mask.nc file was created by running
program obs_seq_coverage with the namelist specified in the
single 48hour forecast evaluated every 6
hours example. The
obsdef_mask.txt file was used to mask the input observation sequence files by
program obs_selection and the result was run through
PROGRAM filter with the observations marked as evaluate_only - resulting in a file called
obs_seq.forecast.2008060818. This filename could also be put in a file called verify_list.txt.obs_seq_coverage and obs_seq_verify are provided below.&obs_seq_coverage_nml
obs_sequences = ''
obs_sequence_list = 'coverage_list.txt'
obs_of_interest = 'METAR_U_10_METER_WIND'
'METAR_V_10_METER_WIND'
textfile_out = 'obsdef_mask.txt'
netcdf_out = 'obsdef_mask.nc'
calendar = 'Gregorian'
first_analysis = 2008, 6, 8, 18, 0, 0
last_analysis = 2008, 6, 8, 18, 0, 0
forecast_length_days = 2
forecast_length_seconds = 0
verification_interval_seconds = 21600
temporal_coverage_percent = 100.0
lonlim1 = 0.0
lonlim2 = 360.0
latlim1 = -90.0
latlim2 = 90.0
verbose = .true.
/
&obs_seq_verify_nml
obs_sequences = 'obs_seq.forecast.2008060818'
obs_sequence_list = ''
station_template = 'obsdef_mask.nc'
netcdf_out = 'forecast.nc'
obtype_string = 'METAR_U_10_METER_WIND'
print_every = 10000
verbose = .true.
debug = .false.
/
The pertinent information from the obsdef_mask.nc file is summarized (from ncdump -v
experiment_times,analysis,forecast_lead obsdef_mask.nc) as follows:
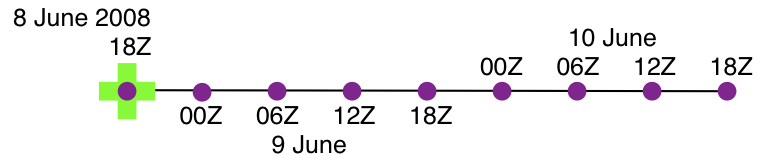
verification_times = 148812.75, 148813, 148813.25, 148813.5, 148813.75,
148814, 148814.25, 148814.5, 148814.75 ;
analysis = 148812.75 ;
forecast_lead = 0, 21600, 43200, 64800, 86400, 108000, 129600, 151200, 172800 ;
There is one analysis time, 9 forecast leads and 9 verification times. The analysis time is the same as the first
verification time. The run-time output of obs_seq_verify and a dump of the resulting netCDF file follows:
[thoar@mirage2 work]$ ./obs_seq_verify |& tee my.verify.log
Starting program obs_seq_verify
Initializing the utilities module.
Trying to log to unit 10
Trying to open file dart_log.out
--------------------------------------
Starting ... at YYYY MM DD HH MM SS =
2011 3 1 10 2 54
Program obs_seq_verify
--------------------------------------
set_nml_output Echo NML values to log file only
Trying to open namelist log dart_log.nml
------------------------------------------------------
-------------- ASSIMILATE_THESE_OBS_TYPES --------------
RADIOSONDE_TEMPERATURE
RADIOSONDE_U_WIND_COMPONENT
RADIOSONDE_V_WIND_COMPONENT
SAT_U_WIND_COMPONENT
SAT_V_WIND_COMPONENT
-------------- EVALUATE_THESE_OBS_TYPES --------------
RADIOSONDE_SPECIFIC_HUMIDITY
------------------------------------------------------
find_ensemble_size: opening obs_seq.forecast.2008060818
location_mod: Ignoring vertical when computing distances; horizontal only
find_ensemble_size: There are 50 ensemble members.
fill_stations: There are 221 stations of interest,
fill_stations: ... and 9 times of interest.
InitNetCDF: METAR_U_10_METER_WIND_forecast.nc is fortran unit 5
obs_seq_verify: opening obs_seq.forecast.2008060818
analysis 1 date is 2008 Jun 08 18:00:00
index 6 is prior ensemble member 1
index 8 is prior ensemble member 2
index 10 is prior ensemble member 3
...
index 100 is prior ensemble member 48
index 102 is prior ensemble member 49
index 104 is prior ensemble member 50
QC index 1 NCEP QC index
QC index 2 DART quality control
Processing obs 10000 of 84691
Processing obs 20000 of 84691
Processing obs 30000 of 84691
Processing obs 40000 of 84691
Processing obs 50000 of 84691
Processing obs 60000 of 84691
Processing obs 70000 of 84691
Processing obs 80000 of 84691
METAR_U_10_METER_WIND dimlen 1 is 9
METAR_U_10_METER_WIND dimlen 2 is 50
METAR_U_10_METER_WIND dimlen 3 is 3
METAR_U_10_METER_WIND dimlen 4 is 1
METAR_U_10_METER_WIND dimlen 5 is 221
METAR_U_10_METER_WIND dimlen 6 is 1
obs_seq_verify: Finished successfully.
--------------------------------------
Finished ... at YYYY MM DD HH MM SS =
2011 3 1 10 3 7
--------------------------------------
[thoar@mirage2 work]$ ncdump -h METAR_U_10_METER_WIND_forecast.nc
netcdf METAR_U_10_METER_WIND_forecast {
dimensions:
analysisT = UNLIMITED ; // (1 currently)
copy = 3 ;
station = 221 ;
level = 14 ;
ensemble = 50 ;
forecast_lead = 9 ;
linelen = 129 ;
nlines = 446 ;
stringlength = 64 ;
location = 3 ;
variables:
char namelist(nlines, linelen) ;
namelist:long_name = "input.nml contents" ;
char CopyMetaData(copy, stringlength) ;
CopyMetaData:long_name = "copy quantity names" ;
double analysisT(analysisT) ;
analysisT:long_name = "time of analysis" ;
analysisT:units = "days since 1601-1-1" ;
analysisT:calendar = "Gregorian" ;
analysisT:missing_value = 0. ;
analysisT:_FillValue = 0. ;
int copy(copy) ;
copy:long_name = "observation copy" ;
copy:note1 = "1 == observation" ;
copy:note2 = "2 == prior" ;
copy:note3 = "3 == observation error variance" ;
copy:explanation = "see CopyMetaData variable" ;
int station(station) ;
station:long_name = "station index" ;
double level(level) ;
level:long_name = "vertical level of observation" ;
int ensemble(ensemble) ;
ensemble:long_name = "ensemble member" ;
int forecast_lead(forecast_lead) ;
forecast_lead:long_name = "forecast lead time" ;
forecast_lead:units = "seconds" ;
double location(station, location) ;
location:description = "location coordinates" ;
location:location_type = "loc3Dsphere" ;
location:long_name = "threed sphere locations: lon, lat, vertical" ;
location:storage_order = "Lon Lat Vertical" ;
location:units = "degrees degrees which_vert" ;
int which_vert(station) ;
which_vert:long_name = "vertical coordinate system code" ;
which_vert:VERTISUNDEF = -2 ;
which_vert:VERTISSURFACE = -1 ;
which_vert:VERTISLEVEL = 1 ;
which_vert:VERTISPRESSURE = 2 ;
which_vert:VERTISHEIGHT = 3 ;
which_vert:VERTISSCALEHEIGHT = 4 ;
double METAR_U_10_METER_WIND(analysisT, station, level, copy, ensemble, forecast_lead) ;
METAR_U_10_METER_WIND:long_name = "forecast variable quantities" ;
METAR_U_10_METER_WIND:missing_value = -888888. ;
METAR_U_10_METER_WIND:_FillValue = -888888. ;
int original_qc(analysisT, station, forecast_lead) ;
original_qc:long_name = "original QC value" ;
original_qc:missing_value = -888888 ;
original_qc:_FillValue = -888888 ;
int dart_qc(analysisT, station, forecast_lead) ;
dart_qc:long_name = "DART QC value" ;
dart_qc:explanation1 = "1 == prior evaluated only" ;
dart_qc:explanation2 = "4 == forward operator failed" ;
dart_qc:missing_value = -888888 ;
dart_qc:_FillValue = -888888 ;
// global attributes:
:creation_date = "YYYY MM DD HH MM SS = 2011 03 01 10 03 00" ;
:source = "$URL$" ;
:revision = "$Revision$" ;
:revdate = "$Date$" ;
:obs_seq_file_001 = "obs_seq.forecast.2008060818" ;
}
[thoar@mirage2 work]$
Discussion
the values of ASSIMILATE_THESE_OBS_TYPES and EVALUATE_THESE_OBS_TYPES are completely irrelevant - again - since
obs_seq_verifyis not actually doing an assimilation.The analysis time from the filename is used to determine which analysis from
obsdef_mask.ncis being considered, and which set of verification times to look for. This is important.The individual
prior ensemble membercopies must be present! Since there are no observations being assimilated, there is no reason to choose the posteriors over the priors.There are 221 locations reporting METAR_U_10_METER_WIND observations at all 9 requested verification times.
The
METAR_U_10_METER_WIND_forecast.ncfile has all the metadata to be able to interpret the METAR_U_10_METER_WIND variable.The analysisT dimension is the netCDF record/unlimited dimension. Should you want to increase the strength of the statistical results, you should be able to trivially
ncrcatmore (compatible) netCDF files together.
References
none - but this seems like a good place to start: The Centre for Australian Weather and Climate Research - Forecast Verification Issues, Methods and FAQ