CLM
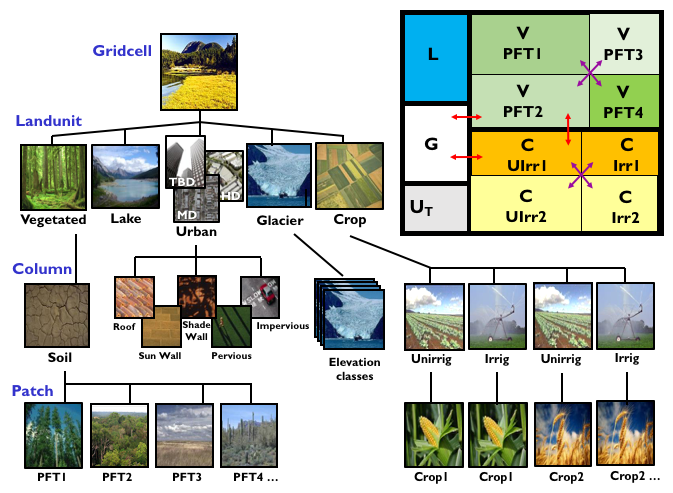 Figure from the CLM5.0 Tech Note. The image is a link to the document.
Figure from the CLM5.0 Tech Note. The image is a link to the document.
Overview
This is the DART interface to the CESM2 Community Land Model. Specifically, CESM release-cesm2.2.0 using CTSM release-cesm2.2.03
This document is most useful if the user has a prior understanding of running CESM and also running CLM-DART. For this reason we strongly recommend the following two steps be completed before reviewing this document:
First, we recommend that you become familiar with running a single or multi-instance experiment in CESM (i.e. a CLM ‘free’ run) before you try to run CLM-DART. The CLM-DART software uses language and concepts that should be familiar to CESM users. The CLM-DART capability is entirely dependent on the multi-instance capability of CESM, first supported in its entirety in CESM1.1.1. Consequently, this version or newer is required to run CLM/DART. The CTSM Documentation has reference material for CLM.
Second, we recommend the user complete the CLM5-DART Tutorial. This tutorial provides the user with 13 steps of hands-on experience for downloading, compiling, editing, executing,and diagnosing a simple CLM5-DART assimilation run. It will provide users with skills to modify the CLM-DART scripts for their own research applications.
DART uses the multi-instance capability of CESM, which means that DART is not responsible for advancing the model. This GREATLY simplifies the traditional DART workflow, but it means CESM has to stop and write out a restart file every time an assimilation is required. The multi-instance capability is relatively new to CESM and we are in close collaboration with the CESM developers to make using DART with CESM as easy as possible. See the SourceMods section for a desciption of the changes to the CLM source code that are useful in a data assimilation context.
CLM is a rapidly-moving target and DART is developed and maintained by a small group of people. Consequently, we have focused on supporting released versions of CLM. This documentation and scripting were tested using the CESM tag release-cesm2.2.0 and CTSM tag release-cesm2.2.03 following the download instructions from https://github.com/ESCOMP/CESM .
CLM-DART has been used to assimilate snow data, soil moisture, leaf area index, biomass, solar-induced fluorescence and more. See the References section below for some examples. The list is by no means complete. Each experiment has extended the capabilities of the CLM-DART system. The observations supported by DART are contained in the obs_def_land_mod.f90, obs_def_tower_mod.f90, and obs_def_COSMOS_mod.f90 modules - each of which may be easily extended. New observation types are usually easily supported. There are many examples of routines to convert from native data formats (netCDF, HDF, csv, etc.) to a DART observation sequence file.
Important Features
Land DA is extremely diverse. The support for Land DA as pertains to CLM-DART has some features that need to be described in some detail.
SourceMods
While we strive to keep DART requirements out of the model code, there are a few SourceMods needed to run DART for CLM from within CESM. DART SourceMods for different versions of CESM are available as part of the DART package at ${dartroot}/models/clm/DART_SourceMods/ where dartroot is the location of your DART installation. It is up to you to either use them ‘as is’ or put them under version control in your CESM source code installation. The DART scripting allows you to specify a directory containing the SourceMods - and then copies those SourceMods to the appropriate CASEROOT directory.
The DART_params.csh file has a large comment block in the section
pertaining to SourceMods.
SourceMod File |
Description |
|---|---|
|
Suppress balance checks for first restart step. This is highly recommended. Many people find it to suppress balance checks for ALL timesteps. |
|
(deprecated) DS199.1 originally had some slightly negative downward radiations that needed to be corrected. |
|
Allows the use of ‘PARVEG’ in a history file. Normally, only ‘PARVEGLN’ is output. |
|
Used to calculate SIF |
|
Used to calculate SIF |
CLM indeterminate values
CLM variables are encoded as rectangular arrays in the netCDF files. However, this means that some variables have space for layers that are unused. Anything with snow layers, for example. CLM has the SNLSNO variable to indicate which snow layers are active. The unused layers may not have the _FillValue value, but can have ‘indeterminate’ values. The PROGRAM clm_to_dart must be run to convert these indeterminate values to _FillValue to be interpreted correctly by DART. After the assimilation is complete, the PROGRAM dart_to_clm must be called to replace the _FillValue with whatever is originally in that slot. This approach preserves the ‘indeterminate’ value for unused snow layers and prevents DART from adjusting the value during the filter step. If the surface snow layer has a trace of snow this is considered an active snow layer, and we allow DART to adjust this value. See the Discussion of Indeterminate Values section of PROGRAM clm_to_dart for more details.
Model Interpolate - The Forward Operator
Since the subgridscale components of CLM have no explicit location associated with them, the location of every component in the gridcell is the same as the gridcell itself. The DART forward operators fundamentally rely on interpolating the model state to some arbitrary location. At present, the best we can do is to create an area-weighted average of all components in the gridcell. This is sub-optimal because it introduces representation mismatch between the grid cell and observation spatial resolution. A nice project would be to use a lookup table for the observation location to determine the dominant PFT (or relevant metadata) at that location and only average the PFTs specifically associated with the observation within the gridcell. This will allow the forward operator to be more accurate and might have a discernable impact on the regression relationship (i.e. ensemble covariance) between the variables in the DART state vector.
The model_interpolate function in DART achieves efficiency by interpolating all the ensemble members at the same time. This gives rise to some challenging problems when interpolating values for variables with with changing numbers of active layers. For example, some ensemble members may only have 2 active snow layers, some may have 3. This is an untenable situation when asked for the snow temperature or water content in layer 3, for example. Consequently - model_interpolate will fail and return an error code - the forward operator will fail - and the observation is rejected and the DART QC is marked as such. Be aware.
Localization
Localization is the term used to restrict the portion of the state to the portion believed to be related to the observation. Most often, this is a spatial argument but it does not need to be restricted to that. In some way, even the selection of the CLM variables to include in the DART state is a de-facto localization. Since CLM has such a rich description of land unit types: urban columns, glaciers, lakes, etc. it is also possible (and probably desirable) to explicitly declare some columns and/or PFTs to be unaffected by the assimilation - i.e., we declare that soil moisture observations should not impact urban columns or deep lakes or … The get_close_state() function employs a routine to explicitly declare what subgridscale components are allowed to be modified by the assimilation. This routine can easily be customized to suit your purpose. The code segment below should make this clear.
! Determine if state_index is a variable from a column (or whatever is of interest).
! Determine what dimension is of interest, need to know to index into
! cols1d_ityplun(ncolumn) array (for example).
RELATEDLOOP: do jdim = 1, get_num_dims(dom_id, var_id)
dimension_name = get_dim_name(dom_id, var_id, jdim)
select case ( trim(dimension_name) )
case ("gridcell","lon","lat")
related = .true.
case ("lndgrid")
related = .true.
case ("landunit")
if ( land1d_ityplun(indices(jdim)) == ilun_vegetated_or_bare_soil ) related = .true.
if ( land1d_ityplun(indices(jdim)) == ilun_crop ) related = .true.
case ("column")
if ( cols1d_ityplun(indices(jdim)) == icol_vegetated_or_bare_soil ) related = .true.
if ( cols1d_ityplun(indices(jdim)) == icol_crop ) related = .true.
case ("pft")
related = .true.
case default
end select
! Since variables can use only one of these dimensions,
! there is no need to check the other dimensions.
if (related) exit RELATEDLOOP
enddo RELATEDLOOP
Snow Data Assimilation
The prognostic variables for snow (i.e. the ones that impact the forecast) are the ones that have layers. The snow observations are typically without explicit depths and are essentially column-integrated quantities like snow water equivalent (SWE - CLM variable H2OSNO) or snow depth (CLM variable SNOW_DEPTH). These CLM diagnostic variables simplify the forward operator and have been part of the DART state. However, updating the prognostic variables (T_SOISNO, H2OSOI_LIQ, H2OSOI_ICE, DZSNO, ZSNO, ZISNO) through their ensemble covariance with the update to H2OSNO, for example will generally not result in a posterior SWE (calculated from the prognostic variables) that matches the posterior SWE in H2OSNO.
In order to address this challenge, a snow repartitition function has been
created in dart_to_clm that redistributes the posterior SWE into
the appropriate prognostic variables. This guarantees that the posterior
SWE of the prognostic snow variables matches the posterior SWE in H2OSNO.
When snow related variables are being updated within an assimilation it is
recommended to invoke this repartitioning function by setting the namelist
option ``repartition_swe = 1 or 2`` within ``&dart_to_clm_nml``. See the
PROGRAM dart_to_clm for more details describing the repartitioning function including
guidance on how to set up a case that repartitions snow. Note that we have not
attempted to include any of the snow property variables most important to controlling
albedo (eg. grain radius, carbon, dust) within the DART state. To what extent adjusting
mass and dimensional properties of snow layers indirectly influences the
albedo properties is an active scientific question. See the PROGRAM dart_to_clm
for more details on how to implement repartition_swe if conserving albedo
is important for your application.
The snow formulation in CLM is complex. Reducing the amount of snow through assimilation is well-defined. Creating snow when there is none is a limited capability in CLM-DART. If snow exists for a subset of ensemble members at a given location, then it is possible to adjust ensemble members with a value of zero to a non-zero value. On the other hand, if all ensemble members do not have snow, or at least one member has a FillValue, the statistical assumptions for ensemble data assimilation are not valid and the snow variables remain at zero. The best method would be to alter the amount of snow from the forcing file and let CLM manage the snow. This is beyond the scope of CLM-DART. We have thought that if one member does not have snow - maybe we should just use the values from some other member - but when does that stop being acceptable? 10 ensemble members? 20? The distributions become multimodal, and the logical end result is that you could wind up using 1 ensemble member to declare the snow for all the remaining members. That seems like a bad idea.
Similar logic applies to the variables related to plant growth. If the LAI observations indicates there should be something growing and nothing has sprouted yet, DART does nothing to the variables.
Configuring an Experiment
Our notes on how to set up, configure, build, and run CESM for an assimilation experiment evolved into scripts. These scripts are not intended to be a ‘black box’; you will have to read and understand them and modify them to your own purpose. They are heavily commented – in keeping with their origins as a set of notes. If you would like to offer suggestions on how to improve those notes - please send them to dart@ucar.edu - we’d love to hear them.
shell_scripts/cesm2_2 |
Description |
|---|---|
|
The script to run first. This configures and builds a very simple single-instance CLM experiment. This is intended to check if CLM runs on your machine. It has nothing to do with DART. There are no DART requirements in it. |
|
Resource file for use when running CLM and DART. This file has all the configuration items needed and will be copied into the CASEROOT directory to be used during an experiment. Other setup scripts within this table require the parameter values defined in this file. |
|
This script takes the single (spun-up) CLM state supplied with the compset and forecasts an ensemble of these. Each ensemble member uses a unique data atmosphere stream file. After some time, the ensemble of CLM states have enough diversity to be a useful initial ensemble for an assimilation experiment. In this configuration, no observations are required, and no DART exectuables are involved. |
|
Given an ensemble of CLM states, advance the ensemble using unique DATM stream files for each ensemble member. This starts from a CESM ‘hybrid’ run-type, so the initial staging of the ensemble is required (and performed by this script). In this configuration, no observations are required, and no DART exectuables are involved. |
|
Takes a single instance from a spun-up ensemble and
advances CLM in 24-hour segments. If that works, the
setup can be extended to run |
|
Runs a multi-instance CLM experiment and can be used to
perform an assimilation.
CLM advances in 24-hour segments. If that works, the
setup can be extended to run |
|
Augments a CESM case with the bits and pieces required to
run DART. When either |
|
This script is invoked by CESM when specified by the
DATA_ASSIMILATION_SCRIPT resource. This resource is
specified by |
|
This script is invoked by CESM when specified by the
DATA_ASSIMILATION_SCRIPT resource. This resource is
specified by |
Declaring the Variables in the DART State
The DART state vector is constructed in a very flexible manner. A namelist is used to relate the netCDF variable name, the netCDF file type [restart, (XY) history, or vector history] with a DART QUANTITY. Including variables from an ‘XY’ CLM history file allows the inclusion of diagnostic variables that can speed up the forward observation operators if gridcell averages are appropriate.
It is also possible to read some variables from the restart file, and some from a ‘vector-based’ history file that has the same structure (gridcell/landunit/column/pft) as the restart file - but may be temporal averages instead of instantaneous quantities. Care must be taken to assign the proper DART QUANTITY to the variables. Any variable in the DART state is updated, but the forward operator looks for specific QUANTITIES. If you want to use the vector-based history file for the forward operator - make sure you declare it to be of the QUANTITY used by the forward operator code.
The namelist specification of what goes into the DART state vector includes the ability to specify if the quantity should have a lower bound, upper bound, or both, what file the variable should be read from, and if the variable should be modified by the assimilation or not. Make sure you read the Inflation section to fully understand what happens when you designate a varible ‘NO_COPY_BACK’.
Attention
It is important to know that the variables in the DART diagnostic files
preassim, postassim, analysis, and output will contain
the unbounded versions of ALL the variables specified in clm_variables.
Only the files specified in the filter_nml:output_state_file_list
will have the ‘clamped’ values.
The example input.nml model_nml demonstrates how to construct the
DART state vector. The following table explains in detail each entry
for clm_variables:
Column |
Description |
|---|---|
1 |
The CLM variable name as it appears in the CLM netCDF file. |
2 |
The corresponding DART QUANTITY. |
3 |
Minimum value of the posterior.
If set to ‘NA’ there is no minimum value.
The DART diagnostic files will not reflect this value, but
the file used to restart CLM will.
|
4 |
Maximum value of the posterior.
If set to ‘NA’ there is no maximum value.
The DART diagnostic files will not reflect this value, but
the file used to restart CLM will.
|
5 |
Specifies which file should be used to obtain the variable.
'restart' => clm_restart_filename'history' => clm_history_filename'vector' => clm_vector_history_filename |
6 |
Should
filter update the variable in the specified file.'UPDATE' => the variable is updated.'NO_COPY_BACK' => the variable remains unchanged. |
The following are only meant to be examples - they are not scientifically validated. Some of these that are UPDATED are probably diagnostic quantities, Some of these that should be updated may be marked NO_COPY_BACK. This list is by no means complete.
clm_variables = 'leafc', 'QTY_LEAF_CARBON', '0.0', 'NA', 'restart' , 'UPDATE',
'frac_sno', 'QTY_SNOWCOVER_FRAC', '0.0', '1.', 'restart' , 'UPDATE',
'SNOW_DEPTH', 'QTY_SNOW_THICKNESS', '0.0', 'NA', 'restart' , 'NO_COPY_BACK',
'H2OSOI_LIQ', 'QTY_SOIL_LIQUID_WATER', '0.0', 'NA', 'restart' , 'UPDATE',
'H2OSOI_ICE', 'QTY_SOIL_ICE', '0.0', 'NA', 'restart' , 'UPDATE',
'T_SOISNO', 'QTY_TEMPERATURE', '0.0', 'NA', 'restart' , 'UPDATE',
'livestemc', 'QTY_LIVE_STEM_CARBON', '0.0', 'NA', 'restart' , 'UPDATE',
'deadstemc', 'QTY_DEAD_STEM_CARBON', '0.0', 'NA', 'restart' , 'UPDATE',
'NEP', 'QTY_NET_CARBON_PRODUCTION', 'NA' , 'NA', 'history' , 'NO_COPY_BACK',
'H2OSOI', 'QTY_SOIL_MOISTURE', '0.0', 'NA', 'history' , 'NO_COPY_BACK',
'SMINN_vr', 'QTY_SOIL_MINERAL_NITROGEN', '0.0', 'NA', 'history' , 'NO_COPY_BACK',
'LITR1N_vr', 'QTY_NITROGEN', '0.0', 'NA', 'history' , 'NO_COPY_BACK',
'TSOI', 'QTY_SOIL_TEMPERATURE', 'NA' , 'NA', 'history' , 'NO_COPY_BACK',
'FSDSVDLN', 'QTY_PAR_DIRECT', '0.0', 'NA', 'history' , 'NO_COPY_BACK',
'FSDSVILN', 'QTY_PAR_DIFFUSE', '0.0', 'NA', 'history' , 'NO_COPY_BACK',
'PARVEGLN', 'QTY_ABSORBED_PAR', '0.0', 'NA', 'history' , 'NO_COPY_BACK',
'NEE', 'QTY_NET_CARBON_FLUX', 'NA' , 'NA', 'vector' , 'NO_COPY_BACK',
'H2OSNO', 'QTY_SNOW_WATER', '0.0', 'NA', 'vector' , 'NO_COPY_BACK',
'TLAI', 'QTY_LEAF_AREA_INDEX', '0.0', 'NA', 'vector' , 'NO_COPY_BACK',
'TWS', 'QTY_TOTAL_WATER_STORAGE', 'NA' , 'NA', 'vector' , 'NO_COPY_BACK',
'SOILC_vr', 'QTY_SOIL_CARBON', '0.0', 'NA', 'vector' , 'NO_COPY_BACK',
'SOIL1N_vr', 'QTY_SOIL_NITROGEN', '0.0', 'NA', 'vector' , 'NO_COPY_BACK',
'SMP', 'QTY_SOIL_MATRIC_POTENTIAL', '0.0', 'NA', 'vector' , 'NO_COPY_BACK'
/
Only the first variable for a DART QUANTITY in the clm_variables list will be used for the forward observation operator. The following is perfectly legal:
clm_variables = 'LAIP_VALUE', 'QTY_LEAF_AREA_INDEX', 'NA', 'NA', 'restart' , 'UPDATE',
'tlai', 'QTY_LEAF_AREA_INDEX', 'NA', 'NA', 'restart' , 'UPDATE',
'elai', 'QTY_LEAF_AREA_INDEX', 'NA', 'NA', 'restart' , 'UPDATE',
'ELAI', 'QTY_LEAF_AREA_INDEX', 'NA', 'NA', 'history' , 'NO_COPY_BACK',
'LAISHA', 'QTY_LEAF_AREA_INDEX', 'NA', 'NA', 'history' , 'NO_COPY_BACK',
'LAISUN', 'QTY_LEAF_AREA_INDEX', 'NA', 'NA', 'history' , 'NO_COPY_BACK',
'TLAI', 'QTY_LEAF_AREA_INDEX', 'NA', 'NA', 'history' , 'NO_COPY_BACK',
'TLAI', 'QTY_LEAF_AREA_INDEX', 'NA', 'NA', 'vector' , 'NO_COPY_BACK'
/
however, only LAIP_VALUE will be used to calculate the LAI when an observation of LAI is encountered. All (the other LAI) variables in the DART state will be modified by the assimilation based on the relationship of LAIP_VALUE and the observation. It is possible that several clm variables could serve as the input for the forward operator, however, in practice, the user should choose the variable that best matches the observation (temporal/spatial resolution, units etc), to help limit the complexity of the forward operator.
Inflation
Inflation has been shown to be quite useful in our experience of DA with CLM and DART. The model is strongly influenced by the atmospheric forcing and will cause the CLM ensemble to relax to a state consistent with the forcing when the assimilation stops. Depending on the forecast length between assimilations, and sometimes just to restore the variance lost during an assimilation, inflation should be used.
The ‘NO_COPY_BACK’ designation has some side effects when it
comes to state-space inflation (inf_flavor 2,4 or 5 -
‘VARYING_SS_INFLATION’,’RELAXATION_TO_PRIOR_SPREAD’,
or ‘ENHANCED_SS_INFLATION’ - respectively). State-space inflation
requires an inflation value for everything in the DART state.
If the variable has been designated as ‘NO_COPY_BACK’
the DART write routine (when called from filter) simply
skips the variable and nothing is written.
This is a problem for inflation files that need to adapt.
The solution is to run
PROGRAM fill_inflation_restart
to create an initial inflation file with inflation values of 1.0 (i.e.
no inflation). fill_inflation_restart has been specially designed
to output inflation values for every variable in the DART state.
The idea is to copy the input inflation file to the output inflation
file name before each assimilation cycle. No new values will be written
for the variables designated ‘NO_COPY_BACK’, the original values will persist.
It remains a scientific question as to whether or not this is the right thing to do! The ‘NO_COPY_BACK’ mechanism was initially intended to simply avoid writing variables that did not impact the next model forecast. Since inflation is a powerful mechanism to overcome observation-model bias, it might be perfectly warranted to ‘UPDATE’ these diagnostic variables. Be warned, if you do ‘UPDATE’ the diagnostic variables, you may want to create copies of the prior so you explore exactly what happens during an assimilation.
If the filter namelist specifies the use of inflation, the assimilate.csh
script is configured to run fill_inflation_restart on the first assimilation cycle.
The inflation filenames are put in a pointer file which is continually updated
as the experiment progresses.
Attention
It is recommended to apply no inflation during the first assimilation step. In other
words within input.nml and namelist &fill_inflation_restart_nml
set prior_inf_mean = 1.00 and post_inf_mean = 1.00. Otherwise, a spatially
uniform inflation will be applied to the entire spatial domain of the assimilation
which can make CLM unstable. In general, inflation is intended to account for biases
between the observation and model-estimated observation, as well as to restore ensemble
spread after an observation has been assimilated.
Namelist
Namelists start with an ampersand ‘&’ and terminate with a slash ‘/’. Character strings that contain a ‘/’ must be enclosed in quotes to prevent them from prematurely terminating the namelist. These are the defaults:
&model_nml
clm_restart_filename = 'clm_restart.nc',
clm_history_filename = 'clm_history.nc',
clm_vector_history_filename = 'clm_vector_history.nc',
output_state_vector = .false.,
assimilation_period_days = 2,
assimilation_period_seconds = 0,
model_perturbation_amplitude = 0.2,
calendar = 'Gregorian',
debug = 0
clm_variables = 'frac_sno', 'QTY_SNOWCOVER_FRAC', 'NA' , 'NA', 'restart' , 'NO_COPY_BACK',
'H2OSNO', 'QTY_SNOW_WATER', '0.0', 'NA', 'restart' , 'UPDATE',
'H2OSOI_LIQ', 'QTY_SOIL_MOISTURE', '0.0', 'NA', 'restart' , 'UPDATE',
'H2OSOI_ICE', 'QTY_ICE', '0.0', 'NA', 'restart' , 'UPDATE',
'T_SOISNO', 'QTY_SOIL_TEMPERATURE', 'NA' , 'NA', 'restart' , 'UPDATE',
'SNOWDP', 'QTY_SNOW_THICKNESS', 'NA' , 'NA', 'restart' , 'UPDATE',
'LAIP_VALUE', 'QTY_LEAF_AREA_INDEX', 'NA' , 'NA', 'restart' , 'NO_COPY_BACK',
'cpool', 'QTY_CARBON', '0.0', 'NA', 'restart' , 'UPDATE',
'frootc', 'QTY_ROOT_CARBON', '0.0', 'NA', 'restart' , 'UPDATE',
'leafc', 'QTY_LEAF_CARBON', '0.0', 'NA', 'restart' , 'UPDATE',
'leafn', 'QTY_LEAF_NITROGEN', '0.0', 'NA', 'restart' , 'UPDATE',
'NEP', 'QTY_NET_CARBON_PRODUCTION', 'NA' , 'NA', 'history' , 'NO_COPY_BACK',
'TV', 'QTY_VEGETATION_TEMPERATURE', 'NA' , 'NA', 'vector' , 'NO_COPY_BACK',
'RH2M_R', 'QTY_SPECIFIC_HUMIDITY', 'NA' , 'NA', 'vector' , 'NO_COPY_BACK',
'PBOT', 'QTY_SURFACE_PRESSURE', 'NA' , 'NA', 'vector' , 'NO_COPY_BACK',
'TBOT', 'QTY_TEMPERATURE', 'NA' , 'NA', 'vector' , 'NO_COPY_BACK'
/
Item |
Type |
Description |
|---|---|---|
clm_restart_filename |
character(len=256) |
this is the filename of the CLM restart file. The DART scripts resolve linking the specific CLM restart file to this generic name. This file provides the elements used to make up the DART state vector. The variables are in their original landunit, column, and PFT-based representations. |
clm_history_filename |
character(len=256) |
this is the filename of the CLM
|
clm_vector_history_filename |
character(len=256) |
this is the filename of a second CLM
history file. The DART scripts
resolve linking the specific CLM
history file to this generic name.
The default setup scripts actually
create 3 separate CLM history files,
the |
output_state_vector |
logical |
If .true. write state vector as a 1D array to the DART diagnostic output files. If .false. break state vector up into variables before writing to the output files. |
assimilation_period_days,
assimilation_period_seconds
|
integer |
Combined, these specify the width of the assimilation window. The current model time is used as the center time of the assimilation window. All observations in the assimilation window are assimilated. BEWARE: if you put observations that occur before the beginning of the assimilation_period, DART will error out because it cannot move the model ‘back in time’ to process these observations. |
model_perturbation_amplitude |
real(r8) |
Required by the DART interfaces, but not used by CLM. |
calendar |
character(len=32) |
string specifying the calendar to use with DART. The CLM dates will be interpreted with this same calendar. For assimilations with real observations, this should be ‘Gregorian’. |
debug |
integer |
Set to 0 (zero) for minimal output. Successively higher values generate successively more output. Not all values are important, however. It seems I’ve only used values [3,6,7,8]. Go figure. |
clm_variables |
character(:,6) |
Strings that identify the CLM
variables, their DART QUANTITY, the min &
max values, what file to read from,
and whether or not the file should be
updated after the assimilation.
The DART QUANTITY must be one found in
|
Modules used
default_model_mod
distributed_state_mod
ensemble_manager_mod
mpi_utilities_mod
netcdf_utilities_mod
obs_def_utilities_mod
obs_kind_mod
options_mod
state_structure_mod
threed_sphere/location_mod
time_manager_mod
types_mod
utilities_mod
Files
filename |
purpose |
|---|---|
input.nml |
to read the model_mod namelist |
clm_restart.nc |
both read and modified by the CLM model_mod |
clm_history.nc |
read by the CLM model_mod for metadata and possible diagnostic variables. |
clm_vector_history.nc |
read by the CLM model_mod for possible diagnostic variables. |
dart_log.out |
the run-time diagnostic output |
dart_log.nml |
the record of all the namelists actually USED - contains the default values |
Error codes and conditions
Routine |
Message |
Comment |
|---|---|---|
nc_write_model_atts |
Various netCDF-f90 interface error messages |
From one of the netCDF calls in the named routine |
Future plans:
Implement a lookup table that relates the observation location to a dominant PFT or COLUMN so the model_interpolate code can average quantities from similar PFTs or COLUMNs instead of everything in the entire grid cell.
Implement a fast way to get the quantities needed for the calculation of radiative transfer models - needs a whole column of CLM variables, redundant if multiple frequencies are used.
Figure out what to do when one or more of the ensemble members does not have snow/leaves/etc. when the observation indicates there should be. Ditto for removing snow/leaves/etc. when the observation indicates otherwise.
Right now, the soil moisture observation operator is used by the COSMOS code to calculate the expected neutron intensity counts. This is the right idea, however, the COSMOS forward operator uses m3/m3 and the CLM units are kg/m2. I have not checked to see if they are, in fact, identical. This brings up a bigger issue in that the soil moisture observation operator would also be used to calculate whatever a TDT probe or ??? would measure. What units are they in? Can one operator support both?
References
The CTSM Documentation is THE reference for CLM. Below are a list of CLM-DART scientific publications:
Zhang, Y.-F., T. J. Hoar, Z.-L. Yang, J. L. Anderson, A. M. Toure and M. Rodell, 2014: Assimilation of MODIS snow cover through the Data Assimilation Research Testbed and the Community Land Model version 4. Journal of Geophysical Research: Atmospheres, 142 1489-1508, doi:10.1002/2013JD021329
Lin, P., J. Wei, Z. -L. Yang, Y. Zhang, K. Zhang, 2016: Snow data assimilation‐constrained land initialization improves seasonal temperature prediction. Geophysical Research Letters 43 (21), 11,423-11,432 doi:10.1002/2016GL070966
Zhao, L., Z. -L. Yang and T. J. Hoar, 2016: Global soil moisture estimation by assimilating AMSR-E brightness temperatures in a coupled CLM4-RTM-DART system. Journal of Hydrometeorology, 17, 2431-2454, doi:10.1175/JHM-D-15-0218.1
Kwon, Y., Z. -L. Yang, T. J. Hoar and A. M. Toure, 2017: Improving the radiance assimilation performance in estimating snow water storage across snow and land-cover types in North America. Journal of Hydrometeorology, 18, 651-668, doi:10.1175/JHM-D-16-0102.1
Fox, A. M., Hoar, T. J., Anderson, J. L., Arellano, A. F., Smith, W. K., Litvak, M. E., et al., 2018: Evaluation of a data assimilation system for land surface models using CLM4.5. Journal of Advances in Modeling Earth Systems, 10, 2471–2494, doi.org/10.1029/2018MS001362
Ling, X. L., Fu, C. B., Yang, Z. L., & Guo, W. D., 2019: Comparison of different sequential assimilation algorithms for satellite-derived leaf area index using the Data Assimilation Research Testbed (version Lanai). Geoscientific Model Development, 12(7), 3119-3133. doi.org/10.5194/gmd-12-3119-2019
Bian, Q., Xu, Z., Zhao, L., Zhang, Y. F., Zheng, H., Shi, C., … & Yang, Z. L., 2019: Evaluation and intercomparison of multiple snow water equivalent products over the Tibetan Plateau. Journal of Hydrometeorology, 20(10), 2043-2055. doi.org/10.1175/JHM-D-19-0011.1
Raczka, B., Hoar T.J., Duarte H.F., Fox A.M., Anderson J.L., Bowling D.R., & Lin J.C., 2021 Improving CLM5.0 Biomass and Carbon Exchange across the Western US Using a Data Assimilation System. Journal of Advances in Modeling Earth Systems, doi.org/10.1029/2020MS002421
Attention
The remainder of the document describes the deprecated scripts in the shell_scripts/cesm1_x directory - for reference only. These scripts will not work with CESM2.
Script |
Description |
|---|---|
|
runs a single instance of CLM to harvest synthetic observations for an OSSE or “perfect model” experiment. It requires a single CLM state from a previous experiment and uses a specified DATM stream for forcing. This parallels an assimilation experiment in that in the multi-instance setting each CLM instance may use (should use?) a unique DATM forcing. This script has almost nothing to do with DART. There is one (trivial) section that records some configuration information in the DART setup script, but that’s about it. This script should initially be run without DART to ensure a working CESM environment. As of (V7195) 3 October 2014, this script demonstrates how to create ‘vector’-based CLM history files (which requires a bugfix) and has an option to use a bugfixed snow grain-size code. http://bugs.cgd.ucar.edu/show_bug.cgi?id=1730 http://bugs.cgd.ucar.edu/show_bug.cgi?id=1934 |
|
Is functionally identical to |
|
runs a multi-instance CLM experiment and can be used to perform a free run or ‘open loop’ experiment. By default, each CLM instance uses a unique DATM forcing. This script also has almost nothing to do with DART. There is one (trivial) section that records some configuration information in the DART setup script, but that’s about it. This script should initially be run without DART to ensure a working CESM. As of (V7195) 3 October 2014, this script demonstrates how to create ‘vector’-based CLM history files (which requires a bugfix) and has an option to use a bugfixed snow grain-size code. http://bugs.cgd.ucar.edu/show_bug.cgi?id=1730 http://bugs.cgd.ucar.edu/show_bug.cgi?id=1934 |
|
Is functionally identical to |
|
augments a CESM case with the bits and pieces required to
run DART. When either |
In addition to the script above, there are a couple scripts that will either perform an assimilation
(assimilate.csh) or harvest observations for a perfect model experiment
(perfect_model.csh). These scripts are designed to work on several compute
platforms although they require configuration, mainly to indicate the location of the DART observation sequence files on
your system.