MPAS_ATM
Overview
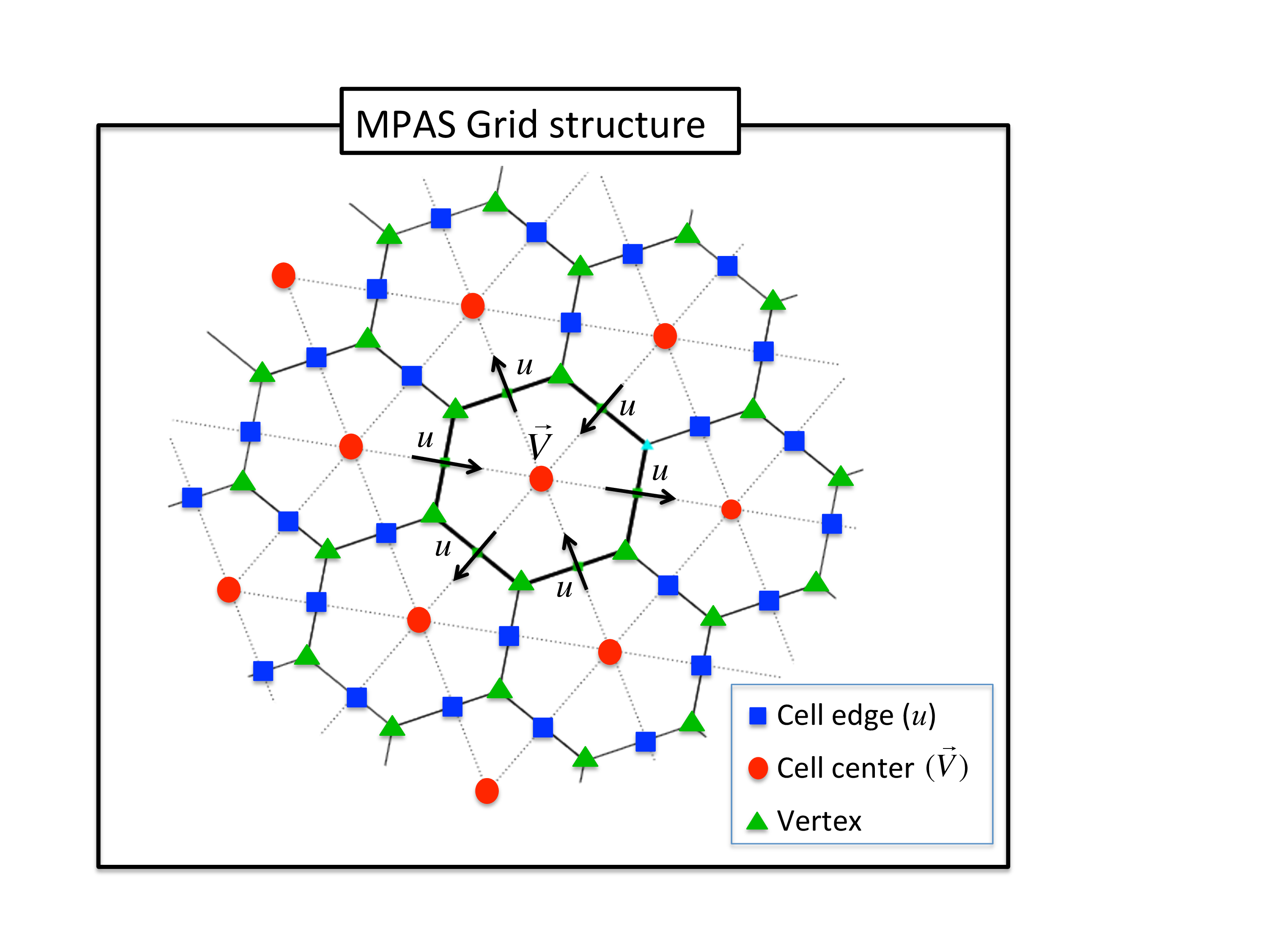
This document describes the DART interface module for the atmospheric component of the Model for Prediction Across Scales MPAS (or briefly, MPAS-ATM) global model, which uses an unstructured Voronoi grid mesh, formally Spherical Centriodal Voronoi Tesselations (SCVTs). This allows for both quasi-uniform discretization of the sphere and local refinement. The MPAS/DART interface was built on the SCVT-dual mesh and does not regrid to regular lat/lon grids. In the C-grid discretization, the normal component of velocity on cell edges is prognosed; zonal and meridional wind components are diagnosed on the cell centers. We provide several options to choose from in the assimilation of wind observations as shown below.
The grid terminology used in MPAS is as shown in the figure below:
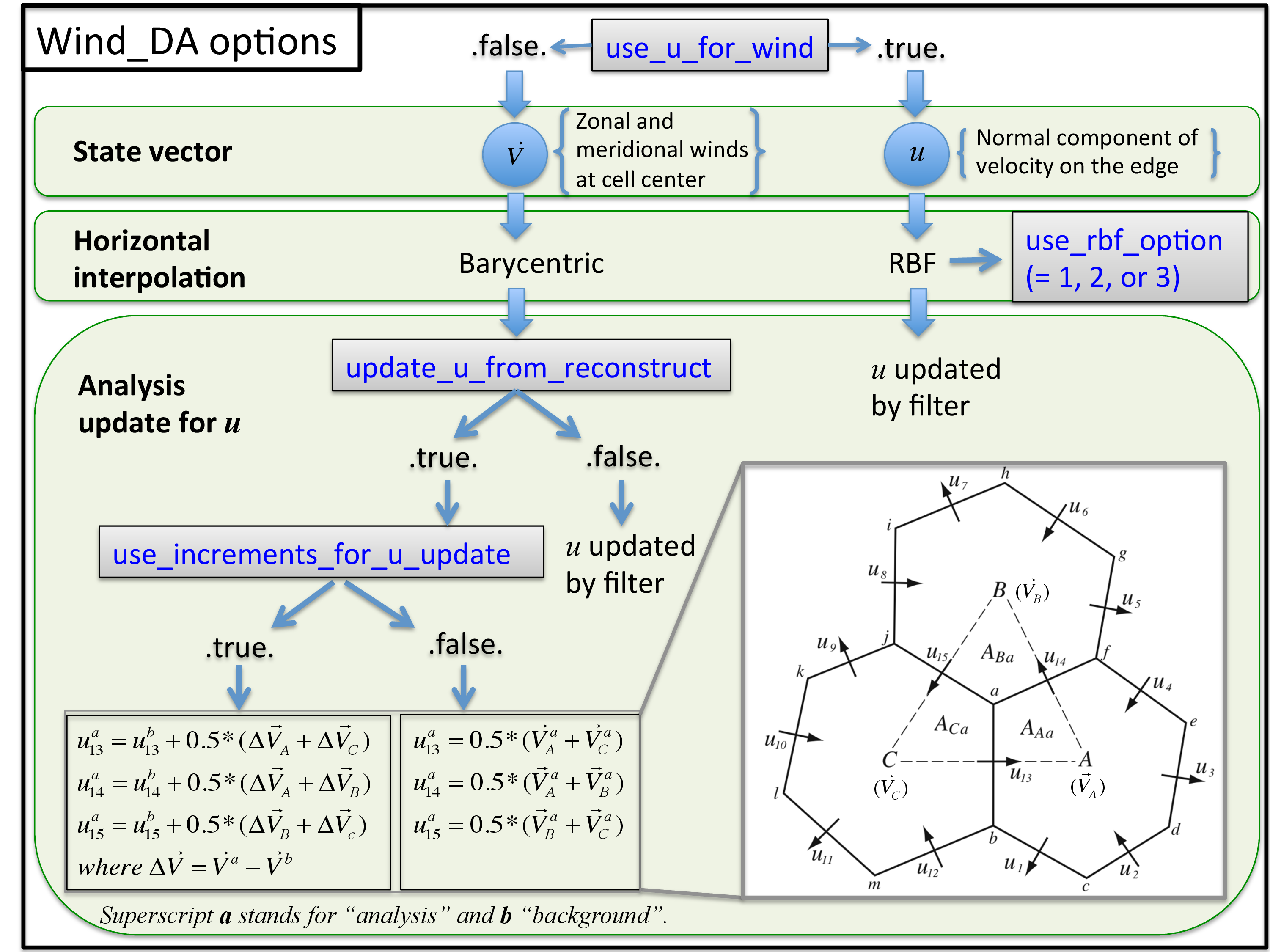
The wind options during a DART assimilation are controlled by combinations of 4 different namelist values. The values determine which fields the forward operator uses to compute expected observation values; how the horizontal interpolation is computed in that forward operator; and how the assimilation increments are applied to update the wind quantities in the state vector. Preliminary results based on real data assimilation experiments indicate that performance is better when the zonal and meridional winds are used as input to the forward operator that uses Barycentric interpolation, and when the prognostic u wind is updated by the incremental method described in the figure below. However there remain scientific questions about how best to handle the wind fields under different situations. Thus we have kept all implemented options available for use in experimental comparisons. See the figure below for a flow-chart representation of how the 4 namelist items interact:
Cycling of MPAS/DART is run in a restart mode. As for all DART experiments,
the overall design for an experiment is this: the DART program filter will
read the initial condition file, the observation sequence file, and the DART
namelist to decide whether or not to advance the MPAS-ATM model. All of the
control of the execution of the MPAS model is done by DART directly. If the
model needs to be advanced, filter makes a call to the shell to execute the
script advance_model.csh, which is ENTIRELY responsible for getting all the
input files, data files, namelists, etc. into a temporary directory, running the
model, and copying the results back to the parent directory (which we call
CENTRALDIR). The whole process hinges on setting the MPAS-ATM model namelist
values such that it is doing a restart for every model advance. Unlike MPAS-ATM
free forecast runs, the forecast step in MPAS/DART requires to set up one more
namelist parameter called config_do_DAcycling = .true. in &restart
section of namelist.input to recouple the state vectors (updated by filter)
with the mass field for the restart mode. For more information, check the
advance_model.csh script in ./shell_scripts/ directory.
Since DART is an ensemble algorithm, there are multiple analysis files for a single analysis time: one for each ensemble member. Because MPAS/DART is run in a restart mode, each member should keep its own MPAS restart file from the previous cycle (rather than having a single template file in CENTRALDIR). Creating the initial ensemble of states is an area of active research.
Namelist
This namelist is read from the file input.nml. Namelists start with an ampersand ‘&’ and terminate with a slash ‘/’. Character strings that contain a ‘/’ must be enclosed in quotes to prevent them from prematurely terminating the namelist.
&model_nml
init_template_filename = 'mpas_init.nc',
vert_localization_coord = 3,
assimilation_period_days = 0,
assimilation_period_seconds = 21600,
model_perturbation_amplitude = 0.0001,
log_p_vert_interp = .true.,
calendar = 'Gregorian',
use_u_for_wind = .false.,
use_rbf_option = 2,
update_u_from_reconstruct = .true.,
use_increments_for_u_update = .true.,
highest_obs_pressure_mb = 100.0,
sfc_elev_max_diff = -1.0,
outside_grid_level_tolerance = -1.0,
extrapolate = .false.,
debug = 0,
/
Item |
Type |
Description |
|---|---|---|
init_template_filename |
character(len=256) [default: ‘mpas_init.nc’] |
The name of the MPAS analysis file to be read and/or written by the DART programs for the state data. |
highest_obs_pressure_mb |
real(r8) [default: 100.0] |
Observations higher than this pressure are ignored. Set to -1.0 to ignore this test. For models with a prescribed top boundary layer, trying to assimilate very high observations results in problems because the model damps out any changes the assimilation tries to make. With adaptive algorithms this results in larger and larger coefficients as the assimilation tries to effect state vector change. |
assimilation_period_days |
integer [default: 0] |
The number of days to advance the model for each assimilation. Even if the model is being advanced outside of the DART filter program, the assimilation period should be set correctly. Only observations with a time within +/- 1/2 this window size will be assimilated. |
assimilation_period_seconds |
integer [default: 21600] |
In addition to
|
vert_localization_coord |
integer [default: 3] |
Vertical coordinate for vertical localization.
|
sfc_elev_max_diff |
real(r8)[default: -1.0] |
If > 0, the maximum difference, in meters, between an observation marked as a ‘surface obs’ as the vertical type (with the surface elevation, in meters, as the numerical vertical location), and the surface elevation as defined by the model. Observations further away from the surface than this threshold are rejected and not assimilated. If the value is negative, this test is skipped. |
log_p_vert_interp |
logical [default: .true.] |
If |
use_u_for_wind |
logical [default: .false.] |
If |
use_rbf_option |
integer [default: 2] |
If |
update_u_from_reconstruct |
logical [default: .true.] |
When zonal and meridional winds at
cell centers are used for the wind
observation operator
( |
use_increments_for_u_update |
logical [default: .true.] |
Only if |
model_perturbation_amplitude |
real(r8) [default: 0.0001] |
The amplitude of random noise to add
when trying to perturb a single state
vector to create an ensemble. Only
used when |
calendar |
character(len=32) [default: ‘Gregorian’] |
Character string specifying the calendar being used by MPAS. |
outside_grid_level_tolerance |
real(r8) [default: -1.0] |
If greater than 0.0, amount of
distance in fractional model levels
that a vertical location can be above
or below the top or bottom of the
grid and still be evaluated without
error. Since extrapolate is not
implemented yet, the value of
|
extrapolate |
logical [default: .false.] |
NOT IMPLEMENTED YET. Vertical
locations equivalant to level 1 or
level N will be used. When this is
implemented, it will do:
If outside_grid_level_tolerance is
greater than 0.0, then control how
values are assigned to locations
where the vertical is exterior to the
grid. If this is |
debug |
integer [default: 0] |
The switch to specify the run-time verbosity.
|
The &mpas_vars_nml namelist within input.nml contains the list of MPAS
variables that make up the DART state vector. The order the items are specified
controls the order of the data in the state vector, so it should not be changed
without regenerating all DART initial condition or restart files. These
variables are directly updated by the filter assimilation.
Any variables whose values cannot exceed a given minimum or maximum can be
listed in mpas_state_bounds. When the data is written back into the MPAS
NetCDF files values outside the allowed range will be detected and changed. Data
inside the DART state vector and data written to the DART diagnostic files will
not go through this test and values may exceed the allowed limits. Note that
changing values at the edges of the distribution means it is no longer
completely gaussian. In practice this technique has worked effectively, but if
the assimilation is continually trying to move the values outside the permitted
range the results may be of poor quality. Examine the diagnostics for these
fields carefully when using bounds to restrict their values.
&mpas_vars_nml
mpas_state_variables = 'theta', 'QTY_POTENTIAL_TEMPERATURE',
'uReconstructZonal', 'QTY_U_WIND_COMPONENT',
'uReconstructMeridional','QTY_V_WIND_COMPONENT',
'qv', 'QTY_VAPOR_MIXING_RATIO',
'qc', 'QTY_CLOUDWATER_MIXING_RATIO',
'surface_pressure', 'QTY_SURFACE_PRESSURE'
mpas_state_bounds = 'qv','0.0','NULL','CLAMP',
'qc','0.0','NULL','CLAMP',
/
Item |
Type |
Description |
|---|---|---|
mpas_vars_nml |
character(len=NF90_MAX_NAME):: dimension(160) |
The table that both specifies which
MPAS-ATM variables will be placed in the
state vector, and also relates those
variables to the corresponding DART kinds.
The first column in each pair must be the
exact NetCDF name of a field in the MPAS
file. The second column in each pair must
be a KIND known to the DART system. See
the |
mpas_state_bounds |
character(len=NF90_MAX_NAME):: dimension(160) |
List only MPAS-ATM variables that must restrict their values to remain between given lower and/or upper bounds. Columns are: NetCDF variable name, min value, max value, and action to take for out-of-range values. Either min or max can have the string ‘NULL’ to indicate no limiting will be done. If the action is ‘CLAMP’ out of range values will be changed to the corresponding bound and execution continues; ‘FAIL’ stops the executable if out of range values are detected. |
Grid Information
As the forward operators use the unstructured grid meshes in MPAS-ATM, the
DART/MPAS interface needs to read static variables related to the grid structure
from the MPAS ATM ‘history’ file (specified in model_analysis_filename).
These variables are used to find the closest cell to an observation point in the
cartesian coordinate (to avoid the polar issues).
integer :: nCells |
the number of cell centers |
integer :: nEdges |
the number of cell edges |
integer :: nVertices |
the number of cell vertices |
integer :: nVertLevels |
the number of vertical levels for mass fields |
integer :: nVertLevelsP1 |
the number of vertical levels for vertical velocity |
integer :: nSoilLevels |
the number of soil levels |
real(r8) :: latCell(:) |
the latitudes of the cell centers [-90,90] |
real(r8) :: lonCell(:) |
the longitudes of the cell centers [0, 360] |
real(r8) :: latEdge(:) |
the latitudes of the edges [-90,90], if edge winds are used. |
real(r8) :: lonEdge(:) |
the longitudes of the edges [0, 360], if edge winds are used. |
real(r8) :: xVertex(:) |
The cartesian location in x-axis of the vertex |
real(r8) :: yVertex(:) |
The cartesian location in y-axis of the vertex |
real(r8) :: zVertex(:) |
The cartesian location in z-axis of the vertex |
real(r8) :: xEdge(:) |
The cartesian location in x-axis of the edge, if edge winds are used. |
real(r8) :: yEdge(:) |
The cartesian location in y-axis of the edge, if edge winds are used. |
real(r8) :: zEdge(:) |
The cartesian location in z-axis of the edge, if edge winds are used. |
real(r8) :: zgrid(:,:) |
geometric height at cell centers (nCells, nVertLevelsP1) |
integer :: CellsOnVertex(:,:) |
list of cell centers defining a triangle |
integer :: edgesOnCell(:,:) |
list of edges on each cell |
integer :: verticesOnCell(:,:) |
list of vertices on each cell |
integer :: edgeNormalVectors(:,:) |
unit direction vectors on the edges (only used if use_u_for_wind = .true.) |
model_mod variable storage
The &mpas_vars_nml within input.nml defines the list of MPAS variables
used to build the DART state vector. Combined with an MPAS analysis file, the
information is used to determine the size of the DART state vector and derive
the metadata. To keep track of what variables are contained in the DART state
vector, an array of a user-defined type called “progvar” is available with the
following components:
type progvartype
private
character(len=NF90_MAX_NAME) :: varname
character(len=NF90_MAX_NAME) :: long_name
character(len=NF90_MAX_NAME) :: units
character(len=NF90_MAX_NAME), dimension(NF90_MAX_VAR_DIMS) :: dimname
integer, dimension(NF90_MAX_VAR_DIMS) :: dimlens
integer :: xtype ! netCDF variable type (NF90_double, etc.)
integer :: numdims ! number of dimensions - excluding TIME
integer :: numvertical ! number of vertical levels in variable
integer :: numcells ! number of cell locations (typically cell centers)
integer :: numedges ! number of edge locations (edges for normal velocity)
logical :: ZonHalf ! vertical coordinate for mass fields (nVertLevels)
integer :: varsize ! variable size (dimlens(1:numdims))
integer :: index1 ! location in dart state vector of first occurrence
integer :: indexN ! location in dart state vector of last occurrence
integer :: dart_kind
character(len=paramname_length) :: kind_string
logical :: clamping ! does variable need to be range-restricted before
real(r8) :: range(2) ! lower and upper bounds for the data range.
logical :: out_of_range_fail ! is out of range fatal if range-checking?
end type progvartype
type(progvartype), dimension(max_state_variables) :: progvar
The variables are simply read from the MPAS analysis file and stored in the DART state vector such that all quantities for one variable are stored contiguously. Within each variable; they are stored vertically-contiguous for each horizontal location. From a storage standpoint, this would be equivalent to a Fortran variable dimensioned x(nVertical,nHorizontal,nVariables). The fastest-varying dimension is vertical, then horizontal, then variable … naturally, the DART state vector is 1D. Each variable is also stored this way in the MPAS analysis file.
Compilation
The DART interface for MPAS-ATM can be compiled with various fortran compilers such as (but not limited to) gfortran, pgf90, and intel. It has been tested on a Mac and NCAR IBM supercomputer (yellowstone).
Note
While MPAS requires the PIO (Parallel IO) and pNetCDF (Parallel NetCDF) libraries, DART uses only the plain NetCDF libraries. If an altered NetCDF library is required by the parallel versions, there may be incompatibilities between the run-time requirements of DART and MPAS. Static linking of one or the other executable, or swapping of modules between executions may be necessary.
Conversions
A Welcome Development
MPAS files no longer need to be converted to DART formatted files, they can be read in directly from a input file list!
Analysis File NetCDF header
The header of an MPAS analysis file is presented below - simply for context. Keep in mind that many variables have been removed for clarity. Also keep in mind that the multi-dimensional arrays listed below have the dimensions reversed from the Fortran convention. Note: the variables marked ‘available in dart’ are available as metadata variables in DART. Just to be perfectly clear, they are not ‘state’.
$ ncdump -h mpas_init.nc
netcdf mpas_analysis {
dimensions:
StrLen = 64 ;
Time = UNLIMITED ; // (1 currently)
nCells = 10242 ; available in DART
nEdges = 30720 ; available in DART
maxEdges = 10 ;
maxEdges2 = 20 ;
nVertices = 20480 ; available in DART
TWO = 2 ;
THREE = 3 ;
vertexDegree = 3 ;
FIFTEEN = 15 ;
TWENTYONE = 21 ;
R3 = 3 ;
nVertLevels = 41 ; available in DART
nVertLevelsP1 = 42 ; available in DART
nMonths = 12 ;
nVertLevelsP2 = 43 ;
nSoilLevels = 4 ; available in DART
variables:
char xtime(Time, StrLen) ; available in DART
double latCell(nCells) ; available in DART
double lonCell(nCells) ; available in DART
double latEdge(nEdges) ; available in DART
double lonEdge(nEdges) ; available in DART
int indexToEdgeID(nEdges) ;
double latVertex(nVertices) ;
double lonVertex(nVertices) ;
double xVertex(nVertices) ; available in DART
double yVertex(nVertices) ; available in DART
double zVertex(nVertices) ; available in DART
double xEdge(nVertices) ; available in DART
double yEdge(nVertices) ; available in DART
double zEdge(nVertices) ; available in DART
int indexToVertexID(nVertices) ;
int cellsOnEdge(nEdges, TWO) ;
int nEdgesOnCell(nCells) ;
int nEdgesOnEdge(nEdges) ;
int edgesOnCell(nCells, maxEdges) ; available in DART
int edgesOnEdge(nEdges, maxEdges2) ;
double weightsOnEdge(nEdges, maxEdges2) ;
double dvEdge(nEdges) ;
double dcEdge(nEdges) ;
double angleEdge(nEdges) ;
double edgeNormalVectors(nEdges, R3) ; available in DART
double cellTangentPlane(nEdges, TWO, R3) ;
int cellsOnCell(nCells, maxEdges) ;
int verticesOnCell(nCells, maxEdges) ; available in DART
int verticesOnEdge(nEdges, TWO) ;
int edgesOnVertex(nVertices, vertexDegree) ;
int cellsOnVertex(nVertices, vertexDegree) ; available in DART
double kiteAreasOnVertex(nVertices, vertexDegree) ;
double rainc(Time, nCells) ;
double cuprec(Time, nCells) ;
double cutop(Time, nCells) ;
double cubot(Time, nCells) ;
double relhum(Time, nCells, nVertLevels) ;
double qsat(Time, nCells, nVertLevels) ;
double graupelnc(Time, nCells) ;
double snownc(Time, nCells) ;
double rainnc(Time, nCells) ;
double graupelncv(Time, nCells) ;
double snowncv(Time, nCells) ;
double rainncv(Time, nCells) ;
double sr(Time, nCells) ;
double surface_temperature(Time, nCells) ;
double surface_pressure(Time, nCells) ;
double coeffs_reconstruct(nCells, maxEdges, R3) ;
double theta_base(Time, nCells, nVertLevels) ;
double rho_base(Time, nCells, nVertLevels) ;
double pressure_base(Time, nCells, nVertLevels) ;
double exner_base(Time, nCells, nVertLevels) ;
double exner(Time, nCells, nVertLevels) ;
double h_divergence(Time, nCells, nVertLevels) ;
double uReconstructMeridional(Time, nCells, nVertLevels) ;
double uReconstructZonal(Time, nCells, nVertLevels) ;
double uReconstructZ(Time, nCells, nVertLevels) ;
double uReconstructY(Time, nCells, nVertLevels) ;
double uReconstructX(Time, nCells, nVertLevels) ;
double pv_cell(Time, nCells, nVertLevels) ;
double pv_vertex(Time, nVertices, nVertLevels) ;
double ke(Time, nCells, nVertLevels) ;
double rho_edge(Time, nEdges, nVertLevels) ;
double pv_edge(Time, nEdges, nVertLevels) ;
double vorticity(Time, nVertices, nVertLevels) ;
double divergence(Time, nCells, nVertLevels) ;
double v(Time, nEdges, nVertLevels) ;
double rh(Time, nCells, nVertLevels) ;
double theta(Time, nCells, nVertLevels) ;
double rho(Time, nCells, nVertLevels) ;
double qv_init(nVertLevels) ;
double t_init(nCells, nVertLevels) ;
double u_init(nVertLevels) ;
double pressure_p(Time, nCells, nVertLevels) ;
double tend_theta(Time, nCells, nVertLevels) ;
double tend_rho(Time, nCells, nVertLevels) ;
double tend_w(Time, nCells, nVertLevelsP1) ;
double tend_u(Time, nEdges, nVertLevels) ;
double qv(Time, nCells, nVertLevels) ;
double qc(Time, nCells, nVertLevels) ;
double qr(Time, nCells, nVertLevels) ;
double qi(Time, nCells, nVertLevels) ;
double qs(Time, nCells, nVertLevels) ;
double qg(Time, nCells, nVertLevels) ;
double tend_qg(Time, nCells, nVertLevels) ;
double tend_qs(Time, nCells, nVertLevels) ;
double tend_qi(Time, nCells, nVertLevels) ;
double tend_qr(Time, nCells, nVertLevels) ;
double tend_qc(Time, nCells, nVertLevels) ;
double tend_qv(Time, nCells, nVertLevels) ;
double qnr(Time, nCells, nVertLevels) ;
double qni(Time, nCells, nVertLevels) ;
double tend_qnr(Time, nCells, nVertLevels) ;
double tend_qni(Time, nCells, nVertLevels) ;
Files
filename |
purpose |
|---|---|
input.nml |
to read the namelist - model_mod_nml and mpas_vars_nml |
mpas_init.nc |
provides model state, and ‘valid_time’ of the model state |
static.nc |
provides grid dimensions |
true_state.nc |
the time-history of the “true” model state from an OSSE |
preassim.nc |
the time-history of the model state before assimilation |
analysis.nc |
the time-history of the model state after assimilation |
dart_log.out [default name] |
the run-time diagnostic output |
dart_log.nml [default name] |
the record of all the namelists actually USED - contains the default values |
References
The Data Assimilation section in the MPAS documentation found at http://mpas-dev.github.io.