Verifying installation
Note
These verification steps require MATLAB®. UCAR Member Institutions have access to institutional licenses for MATLAB, thus we have created verification tools using it.
The Lorenz model is notoriously sensitive to very small changes; in fact, the story of Lorenz discovering this sensitivity is a classic in the annals of the study of chaos, which in turn was instrumental in the development of data assimilation as a field of study. See The Lorenz 63 model: what is it and why should we care? or What is data assimilation? for more information.
This sensitivity is of practical interest for verifying these results. The initial conditions files and observation sequences are provided in ASCII, which is portable across systems, but there may be some machine-specific round-off error in the conversion from ASCII to machine binary. As Lorenz 63 is such a nonlinear model, extremely small differences in the initial conditions may eventually result in noticeably different model trajectories. Even different compiler flags may cause tiny differences that ultimately result in large differences. Your results should start out looking VERY SIMILAR and may diverge with time.
The simplest way to determine if the installation is successful is to
run some of the functions available in DART/diagnostics/matlab/. If
you have yet to download DART, see Downloading DART for
instructions on how to do so. Usually, we launch MATLAB from the
DART/models/lorenz_63/work directory. First, open your terminal and navigate
to the directory where you downloaded DART. Then, execute the following command
in your terminal to navigate to the DART/models/lorenz_63/work directory.
$ cd DART/models/lorenz_63/work
Note
The lines of code that are marked with a $ symbol are commands that
you will execute in your terminal. To execute a command, type it in
exactly as seen, unless specified otherwise, and press enter.
In the case of this Lorenz model, we know the “true” (by definition)
state of the model that is consistent with the observations, which is
generated by running the perfect_model_obs program. We therefore must first
run perfect_model_obs before launching MATLAB and running our diagnostic
scripts. To do so, execute the following three commands in the terminal.
$ ./quickbuild.sh
$ ./perfect_model_obs
$ ./filter
You are now ready to launch MATLAB. To do so, execute the command
MATLABROOT/bin/matlab -nodesktop, and replace MATLABROOT with the
directory that MATLAB is installed in on your computer. By default, MATLAB is
installed in the following locations:
Windows (64-bit):
C:\Program Files\MATLAB\R20XXx (64-bit MATLAB)
C:\Program Files (x86)\MATLAB\R20XXx (32-bit MATLAB)
Windows (32-bit):
C:\Program Files\MATLAB\R20XXx
Linux:
/usr/local/MATLAB/R20XXx
Mac:
/Applications/MATLAB_R20XXx.app
R20XXx must also be replaced with the version of MATLAB that you have
installed. For example, the full command for a Mac user having installed the
latest version of MATLAB would be /Applications/MATLAB_R2022a.app/bin/matlab -nodesktop.
If the corresponding MATLABROOT listed above does not work, you can identify the
correct directory by opening the MATLAB application and executing the command
matlabroot. This will display the directory where MATLAB is installed. It
will have the correct version already incorporated into the MATLABROOT.
Upon successfully launching MATLAB, the MATLAB header will be outputted to your terminal as it is below.
$ MATLABROOT/bin/matlab -nodesktop
< M A T L A B (R) >
Copyright 1984-2022 The MathWorks, Inc.
R2022a Update 1 (9.12.0.1927505) 64-bit (maci64)
April 6, 2022
Now that you have launched MATLAB, you must now use the MATLAB addpath command
to make the DART/diagnostics/matlab/ functions available for execution in any working
directory.
$ addpath ../../../diagnostics/matlab
You are now ready to run the diagnostics. The following MATLAB scripts (plot_total_err
and plot_ens_time_series,) compare the ensemble members with the truth and can
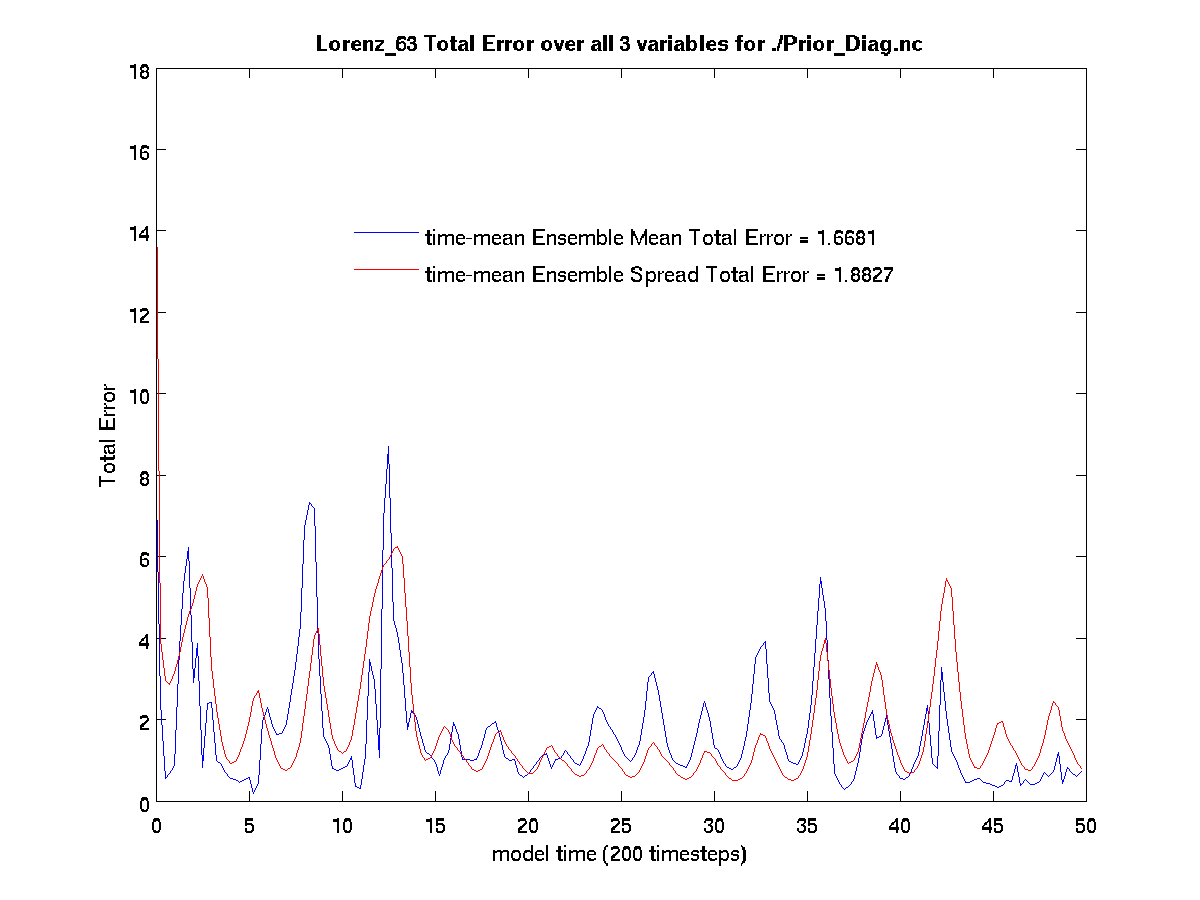
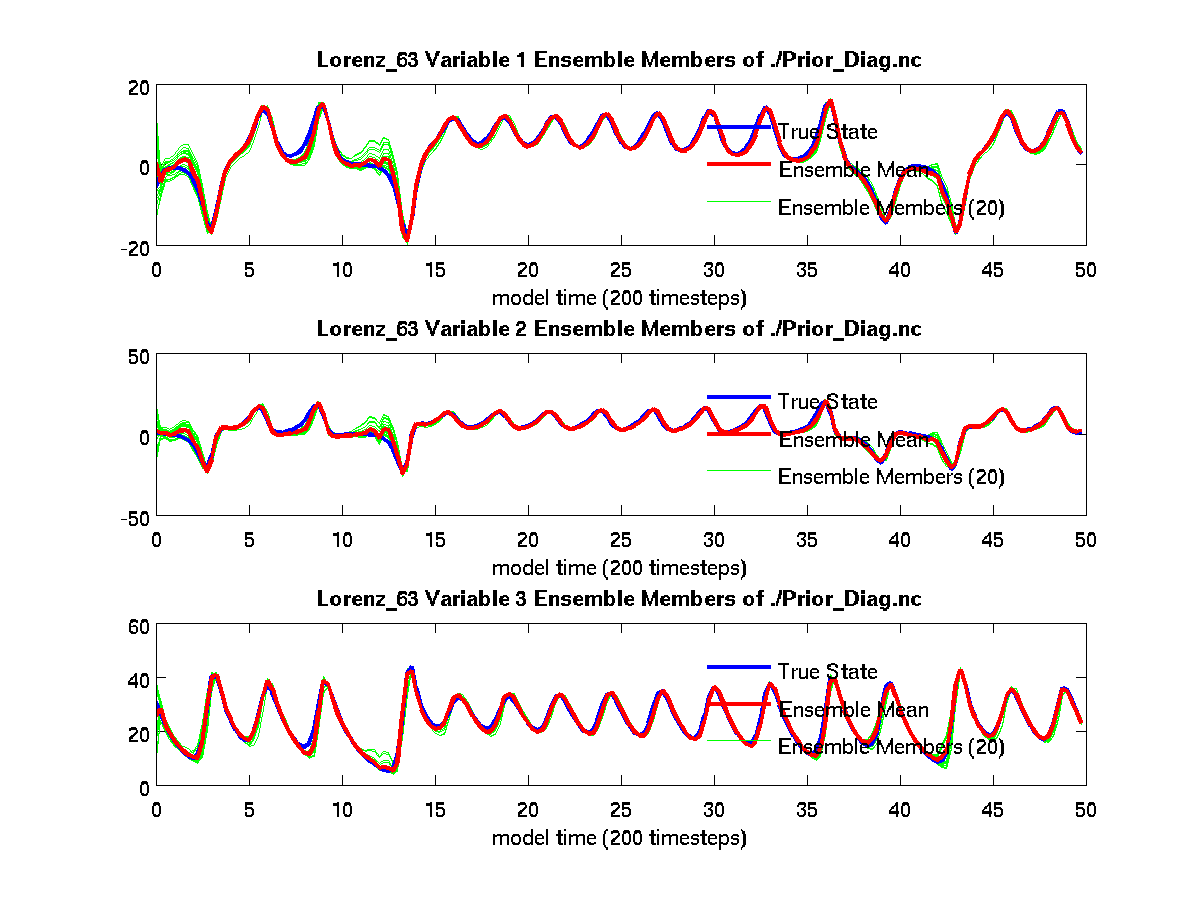
calculate the error in the assimilation. Running plot_total_err and
plot_ens_time_series will produce the graphics as shown below:
Note
When prompted to input file names while running the diagnostics, simply press enter to select the default.
$ plot_total_err
Input name of ensemble trajectory file:
<cr> for preassim.nc
Comparing true_state.nc and
preassim.nc
$ plot_ens_time_series
Input name of ensemble trajectory file:
<cr> for preassim.nc
Comparing true_state.nc and
preassim.nc
Using Variable state IDs 1 2 3
pinfo =
struct with fields:
model: 'Lorenz_63'
def_var: 'state'
num_state_vars: 1
num_copies: 20
num_ens_members: 20
ensemble_indices: [1 2 3 ... 18 19 20]
min_state_var: 1
max_state_var: 3
def_state_vars: [1 2 3]
fname: 'preassim.nc'
truth_file: 'true_state.nc'
diagn_file: 'preassim.nc'
truth_time: [1 200]
diagn_time: [1 200]
vars: {'state'}
time: [200x1 double]
time_series_length: 200
var: 'state'
var_inds: [1 2 3]
From the above plot_ens_time_series graphic, you can see the
individual green ensemble members becoming more constrained with less
spread as time evolves. If your figures look similar to these, you
should feel confident that everything is working as intended. Don’t miss
the opportunity to rotate the “butterfly” plot for that classic chaos
theory experience (perhaps while saying, “life, uh, finds a way”).
Congratulations! You have now successfully configured DART and are ready to begin the next phase of your interaction with DART. You may wish to learn more about:
What is data assimilation? — a brief introduction to ensemble data assimilation. This section includes more information about the Lorenz 63 model and how to configure the
input.nmlfile to play with DA experiments in DART using the Lorenz 63 model.What is DART? — This section includes more information about DART and a basic flow chart of the overall DART workflow.
Note
In the case that the above instructions had one or more issues that either did not work for you as intended or were confusing, please contact the DART software development team at dart@ucar.edu. We value your input to make getting started as smooth as possible for new DART users!